Hypothesis Generation from Literature for Advancing Biological Mechanism Research: A Perspective
New citation alert added.
This alert has been successfully added and will be sent to:
You will be notified whenever a record that you have chosen has been cited.
To manage your alert preferences, click on the button below.

New Citation Alert!
Please log in to your account
Information & Contributors
Bibliometrics & citations, index terms.
Applied computing
Document management and text processing
Life and medical sciences
Bioinformatics
Recommendations
Automated hypothesis generation based on mining scientific literature.
Keeping up with the ever-expanding flow of data and publications is untenable and poses a fundamental bottleneck to scientific progress. Current search technologies typically find many relevant documents, but they do not extract and organize the ...
Research Article: Bioinformatic analysis of molecular network of glucosinolate biosynthesis
Glucosinolates constitute a major group of secondary metabolites in Arabidopsis, which play an important role in plant interaction with pathogens and insects. Advances in glucosinolate research have defined the biosynthetic pathways. However, cross-talk ...
Mining pathway signatures from microarray data and relevant biological knowledge
High-throughput technologies such as DNA microarray are in the process of revolutionising the way modern biological research is being done. Bioinformatics tools are becoming increasingly important to assist biomedical scientists in their quest in ...
Information
Published in.

Association for Computing Machinery
New York, NY, United States
Publication History
Permissions, check for updates, author tags.
- bioinformatics
- biomedical knowledge mining
- machine learning
- Research-article
- Refereed limited
Contributors
Other metrics, bibliometrics, article metrics.
- 0 Total Citations
- 22 Total Downloads
- Downloads (Last 12 months) 22
- Downloads (Last 6 weeks) 1
View Options
Login options.
Check if you have access through your login credentials or your institution to get full access on this article.
Full Access
View options.
View or Download as a PDF file.
View online with eReader .
HTML Format
View this article in HTML Format.
Share this Publication link
Copying failed.
Share on social media
Affiliations, export citations.
- Please download or close your previous search result export first before starting a new bulk export. Preview is not available. By clicking download, a status dialog will open to start the export process. The process may take a few minutes but once it finishes a file will be downloadable from your browser. You may continue to browse the DL while the export process is in progress. Download
- Download citation
- Copy citation
We are preparing your search results for download ...
We will inform you here when the file is ready.
Your file of search results citations is now ready.
Your search export query has expired. Please try again.
- Open access
- Published: 13 June 2024
Dyport: dynamic importance-based biomedical hypothesis generation benchmarking technique
- Ilya Tyagin 1 &
- Ilya Safro 2
BMC Bioinformatics volume 25 , Article number: 213 ( 2024 ) Cite this article
405 Accesses
1 Altmetric
Metrics details
Automated hypothesis generation (HG) focuses on uncovering hidden connections within the extensive information that is publicly available. This domain has become increasingly popular, thanks to modern machine learning algorithms. However, the automated evaluation of HG systems is still an open problem, especially on a larger scale.
This paper presents a novel benchmarking framework Dyport for evaluating biomedical hypothesis generation systems. Utilizing curated datasets, our approach tests these systems under realistic conditions, enhancing the relevance of our evaluations. We integrate knowledge from the curated databases into a dynamic graph, accompanied by a method to quantify discovery importance. This not only assesses hypotheses accuracy but also their potential impact in biomedical research which significantly extends traditional link prediction benchmarks. Applicability of our benchmarking process is demonstrated on several link prediction systems applied on biomedical semantic knowledge graphs. Being flexible, our benchmarking system is designed for broad application in hypothesis generation quality verification, aiming to expand the scope of scientific discovery within the biomedical research community.
Conclusions
Dyport is an open-source benchmarking framework designed for biomedical hypothesis generation systems evaluation, which takes into account knowledge dynamics, semantics and impact. All code and datasets are available at: https://github.com/IlyaTyagin/Dyport .
Peer Review reports
Introduction
Automated hypothesis generation (HG, also known as Literature Based Discovery, LBD) has gone a long way since its establishment in 1986, when Swanson introduced the concept of “Undiscovered Public Knowledge” [ 1 ]. It pertains to the idea that within the public domain, there is a significant abundance of information, allowing for the uncovering of implicit connections among various pieces of information. There are many systems developed throughout the years, which incorporate different reasoning methods: from concept co-occurrence in scientific literature [ 2 , 3 ] to the advanced deep learning-based algorithms and generative models (such as BioGPT [ 4 ] and CBAG [ 5 ]). Examples include but are not limited to probabilistic topic modeling over relevant papers [ 6 ], semantic inference [ 7 ], association rule discovery [ 8 ], latent semantic indexing [ 9 ], semantic knowledge network completion [ 10 ] or human-aware artificial intelligence [ 11 ] to mention just a few. The common thread running through these lines of research is that they are all meant to fill in the gaps between pieces of existing knowledge.
The evaluation of HG is still one of the major problems of these systems, especially when it comes to fully automated large-scale general purpose systems (such as IBM Watson Drug Discovery [ 12 ], AGATHA [ 10 ] or BioGPT [ 4 ]). For these, a massive assessment (that is normal in the machine learning and general AI domains) performed manually by the domain experts is usually not feasible and other methods are required.
One traditional evaluation approach is to make a system “rediscover” some of the landmark findings, similar to what was done in numerous works replicating well-known connections, such as: Fish Oil \(\leftrightarrow\) Raynaud’s Syndrome [ 13 ], Migraine \(\leftrightarrow\) Magnesium [ 13 ] or Alzheimer \(\leftrightarrow\) Estrogen [ 14 ]. This technique is frequently used even in a majority of the recently published papers, despite of its obvious drawbacks, such as very limited number of validation samples and their general obsolesce (some of these connections are over 30 years old). Furthermore, in some of these works, the training set is not carefully chosen to include only the information published prior the discovery of interest which turns the HG goal into the information retrieval task.
Another commonly used technique is based on the time-slicing [ 10 , 15 ], when a system is trained on a subset of data prior to a specified cut-off date and then evaluated on the data from the future. This method addresses the weaknesses of previous approach and can be automated, but it does not immediately answer the question of how significant or impactful the connections are. The lack of this information may lead to deceiving results: many connections, even recently published, are trivial (especially if they are found by the text mining methods) and do not advance the scientific field in a meaningful way.
A related area that faces similar evaluation challenges is Information Extraction (IE), a field crucial to enabling effective HG by identifying and categorizing relevant information in publicly available data sources. Within the realm of biomedical and life sciences IE, there are more targeted, small-scale evaluation protocols such as the BioCreative competitions [ 16 ], where the domain experts provide curated training and test datasets, which allows participants to refine and assess their systems within a controlled environment. While such targeted evaluations as conducted in BioCreative are both crucial and insightful, they inherently lack the scope and scale needed for the evaluation of expansive HG systems.
The aforementioned issues emphasize the critical need for research into effective, scalable evaluation methods in automated hypothesis generation. Our primary interest is in establishing an effective and sustainable benchmark for large-scale, general-purpose automated hypothesis generation systems within the biomedical domain. We seek to identify substantial, non-trivial insights, prioritizing them over mere data volume and ensuring scalability with respect to ever-expanding biocurated knowledge databases. We emphasize the significance of implementing sustainable evaluation strategies, relying on constantly updated datasets reflecting the latest research. Lastly, our efforts are targeted towards distinguishing between hypotheses with significant impact and those with lesser relevance, thus moving beyond trivial generation of hypotheses to ensuring their meaningful contribution to scientific discovery.
Our contribution
We propose a high quality benchmark dataset Dyport for hypothesis prediction systems evaluation. It incorporates information extracted from a number of biocurated databases. We normalize all concepts to the unified format for seamless integration and each connection is supplied with rich metadata, including timestamp information to enable time-slicing.
We introduce an evaluation method for the impact of connections in time-slicing paradigm. It will allow to benchmark HG systems more thoroughly and extensively by assigning an importance weight to every connection over the time. This weight represents the overall impact a connection makes on future discoveries.
We demonstrate the computational results of several prediction algorithms using the proposed benchmark and discuss their performance and quality.
We propose to use our benchmark to evaluate the quality of HG systems. The benchmark is designed to be updated on a yearly basis. Its structure facilitates relatively effortless expansion and reconfiguration by users and developers.
Background and related work
Unfortunately, the evaluation in the hypothesis generation field is often coupled with the systems to evaluate and currently not universally standardized. If one would like to compare the performance of two or more systems, they need to understand their training protocol to instantiate models from scratch and then test them on the same data they used in their experiment.
This problem is well known and there are attempts to provide a universal way to evaluate such systems. For example, OpenBioLink [ 17 ] is designed as a software package for evaluation of link prediction models. It supports time-slicing and contains millions of edges with different quality settings. The authors describe it as “highly challenging” dataset that does not include “trivially predictable” connections, but they do not provide a quantification of difficulty nor range the edges accordingly.
Another attempt to set up a large-scale validation of HG systems was performed in our earlier work [ 18 ]. The proposed methodology is based on the semantic triples extracted from SemMedDB [ 19 ] database and setting up a cut date for training and testing. Triples are converted to pairs by removing the “verb” part from each (subject-verb-object) triple. For the test data, a list of “highly cited” pairs is identified, which is based on the citation counts from SemMedDB, MEDLINE and Semantic Scholar. Only connections occurring in papers published after the cut date and cited over 100 times are considered. It is worth mentioning that this approach is prone to noise (due to SemMedDB text mining methods) and also skewed towards the discoveries published closer to the cut-date, since the citations accumulate over time.
One more aspect of the proposed approach relates to the quantification and detection of scientific novelty. Research efforts range from protein design domain studies [ 20 ] to analyzing scientific publications through their titles [ 21 ] or using manual curation in combination with machine learning [ 22 ]. However, none of these techniques were integrated into a general purpose biomedical evaluation framework, where the novelty would be taken into account.
Currently, Knowledge Graph Embeddings (KGE) are becoming increasingly popular and the hypothesis generation problem can be formulated in terms of link prediction in knowledge graphs. Knowledge Graphs often evaluate the likelihood of a particular connection with the scoring function of choice. For example, TransE [ 23 ] evaluates each sample with the following equation:
where h is the embedding vector of a head entity, r is the embedding vector of relation, t is the embedding vector of a tail entity and \(||\cdot ||\) denotes the L1 or L2 norm.
These days KGE-based models are of interest to the broad scientific community, including researchers in the drug discovery field. Recently they carefully investigated the factors affecting the performance of KGE models [ 24 ] and reviewed biomedical databases related to drug discovery [ 25 ]. These publications, however, do not focus on any temporal information nor attempt to describe the extracted concept associations quantitatively. We also aim to fill in this currently existing gap in our current work.
\(c_i\) —concept in some arbitrary vocabulary;
\(m(\cdot )\) —function that maps a concept \(c_i\) to the subset of corresponding UMLS CUI. The result is denoted by \(m_i =m(c_i)\) . The \(m_i\) is not necessarily a singleton. We will somewhat abuse the notation by denoting \(m_i\) a single or any of the UMLS terms obtained by mapping \(c_i\) to UMLS.
\(m(\cdot ,\cdot )\) —function that maps pairs of \(c_i\) and \(c_j\) into the corresponding set of all possible UMLS pairs \(m_i\) and \(m_j\) . Recall that the mapping of \(c_i\) to UMLS may not be unique. In this case \(|m(c_i,c_j)| = |m(c_i)|\cdot |m(c_j)|\) .
\((m_i, m_j)\) —a pair of UMLS CUIs, which is extracted as a co-occurrence from MEDLINE records. It also represents an edge in network G and is cross-referenced with biocurated databases;
D —set of pairs \((m_i, m_j)\) extracted from biocurated databases;
P —set of pairs \((m_i, m_j)\) extracted from MEDLINE abstracts;
E —set of cross-referenced pairs \((m_i, m_j)\) , such that \(E = D \cap P\) ;
G —dynamic network, containing temporal snapshots \(G_t\) , where t —timestamp (year);
\(\hat{G}_t\) —snapshot of network G for a timestamp t only containing nodes from \(G_{t-1}\) .
The main unit of analysis in HG is a connection between two biomedical concepts, which we also refer to as “pair”, “pairwise interaction” or “edge” (in network science context when we will be discussing semantic networks). These connections can be obtained from two main sources: biomedical databases and scientific texts. Extracting pairs from biomedical databases is done with respect to the nature and content of the database: some of them already contain pairwise interactions, whereas others focus on more complex structures such as pathways which may contain multiple pairwise interactions or motifs (e.g., KEGG [ 26 ]). Extracting pairs from textual data is done via information retrieval methods, such as relation extraction or co-occurrence mining. In this work, we use the abstract-based co-occurrence approach, which is explained later in the paper.
Method in summary
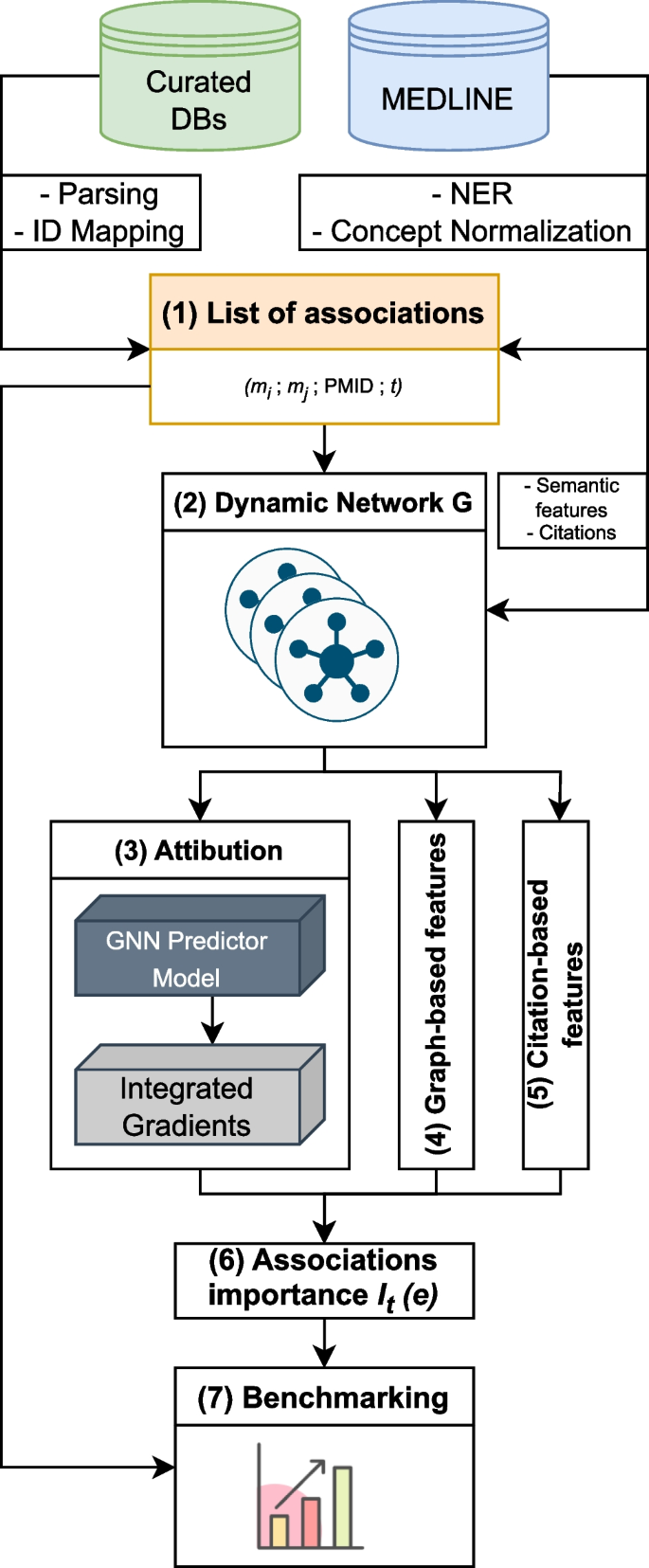
Summary of the HG benchmarking approach. We start with collecting data from Curated DBs and Medline, then process it: records from Curated DBs go through parsing, cleaning and ID mapping, MEDLINE records are fed into SemRep system, which performs NER and concept normalization. After that we obtain a list of UMLS CUI associations with attached PMIDs and timestamps (TS). This data is then used to construct a dynamic network G , which is used to calculate the importance measure I for edges in the network. At the end, edges \(e \in G\) with their corresponding importance scores \(I_t(e)\) are added to the benchmark dataset
The HG benchmarking pipeline is presented in Fig. 1 . The end goal of the pipeline is to provide a way to evaluate any end-to-end hypothesis generation system trained to predict potential pairwise associations between biomedical instances or concepts.
We start with collecting pairwise entity associations from a list of biocurated databases, which we then normalize and represent as pairs of UMLS [ 27 ] terms \((m_i, m_j)\) . The set of these associations is then cross-referenced with scientific abstracts extracted from MEDLINE database, such that for each pair \((m_i, m_j)\) we keep all PubMed identifiers (PMID) that correspond to the paper abstracts in which \(m_i\) and \(m_j\) co-occured. As a result, there is a list of tuples (step 1, Fig. 1 ) \((m_i, m_j, \text {PMID}, t)\) , where t is a timestamp for a given PMID extracted from its metadata. We then split this list into a sequence \(\{E_t\}\) according to the timestamp t . In this work t is taken with a yearly resolution.
Each individual \(E_t\) can be treated as an edgelist, which yields an edge-induced network \(G_t\) constructed from edges \((m_i, m_j) \in E_t\) . It gives us a sequence of networks \(G = \{G_t\}\) (step 2, Fig. 1 ), which is then used to compute the importance of individual associations in \(E_t\) with different methods.
The main goal of importance is to describe each edge from \(E_t\) using additional information. The majority of it comes from the future network snapshot \(G_{t+1}\) , which allows us to track the impact that a particular edge had on the network in the future. The predictive impact is calculated with an attribution technique called Integrated Gradients (IG) (step 3, Fig. 1 ). Structural impact is calculated with graph-based measures (such as centrality) (step 4, Fig. 1 ) and citation impact is calculated with respect to how frequently edges are referenced in the literature after their initial discovery (step 5, Fig. 1 ).
All the obtained scores are then merged together to obtain a ranking \(I_t(e)\) (step 6, Fig. 1 ), where \(e \in E_t\) for all edges from a snapshot \(G_t\) . Finally, this ranking is used to perform stratified evaluation of how well hypothesis generation systems perform in discovering connections with different importance values (step 7, Fig. 1 ).
Databases processing and normalization
We begin by gathering the links and relationships from publicly available databases, curated by domain experts. We ensure that all pairwise concept associations we utilize are from curated sources. For databases like STRING, which compile associations from various channels with differing levels of confidence, we exclusively select associations derived from curated sources.
Ensuring correct correspondence of the same concepts from diverse databases is highly crucial. Therefore, we also conduct mapping of all concepts to UMLS CUI (Concept Unique Identifier). Concepts, which identifiers cannot be mapped to UMLS CUI, are dropped. In our process, we sometimes encounter situations where a concept \(c_{i}\) , may have multiple mappings to UMLS CUIs, i.e., \(|m_i|=k>1\) for \(m_i = m(c_i)\) . To capture these diverse mappings, we use the Cartesian product rule. In this approach, we take the mapping sets for both concepts \(c_{i}\) and \(c_{j}\) , denoted as \(m(c_{i})\) and \(m(c_{j})\) , and generate a new set of pairs encapsulating all possible combinations of these mappings. Essentially, for each original pair \((c_{i}, c_{j})\) , we produce a set of pairs \(m(c_{i}, c_{j})\) such that the cardinality of this new set equals the product of the cardinalities of the individual mappings. Let us say that \(c_i\) has k different UMLS mappings and \(c_j\) has s , then \(|m(c_{1},c_{2})| = |m(c_{1})| \cdot |m(c_{2})| = k\cdot s\) .
In other words, we ensure that every possible mapping of the original pair is accounted for, enabling our system to consider all potential pairwise interactions across all UMLS mappings. To this end, we have collected all pairs of UMLS CUI that are present in different datasets, forming a set D .
Processing MEDLINE records
To match pairwise interactions extracted from biocurated databases to literature, we use records from MEDLINE database with their PubMed identifiers. These records, primarily composed of the titles and abstracts of scientific papers, are each assigned a unique PubMed reference number (PMID). They are also supplemented with rich metadata, which includes information about authors, full-text links (when applicable), and date of publication timestamps indicating when the record became publicly available. We process records with an NLM-developed natural language processing tool SemRep [ 28 ] to perform named entity recognition, concept mapping and normalization. To this end, we obtain a list of UMLS CUI for each MEDLINE record.
Connecting database records with literature
The next step is to form connections between biocurated records and their corresponding mentions in the literature. With UMLS CUIs identified in the previous step, we track the instances where these CUIs are mentioned together within the same scientific abstract. Our method considers the simultaneous appearance of a pair of concepts, denoted as \(m_i\) and \(m_j\) , within a single abstract to represent a co-occurrence. This co-occurrence may indicate a potential relationship between the two concepts within the context of the abstract. All the co-occurring pairs \((m_i, m_j)\) , extracted from MEDLINE abstracts, form the set P .
No specific “significance” score is assigned to these co-occurrences at this point beyond their presence in the same abstract. Subsequently, these pairs are cross-referenced with pairs in biocurated databases. More specifically, for each co-occurrence \((m_i, m_j) \in P\) we check its presence in set D . Pairs not present in both sets D and P are discarded. This forms the set E :
This step validates each co-occurring pair, effectively reducing noise and confirming that each pair holds biological significance. Conversely, E can be described as a set of biologically relevant associations, with each element enriched by contextual information extracted from scientific literature. The procedure is described in [ 29 ] as distant supervised annotation .
Constructing time-sliced graphs
After we find the set of co-occurrences in abstracts extracted from MEDLINE and cross-referenced with pairs in biocurated databases (set E ), we split it based on the timestamps extracted from the abstracts metadata. The timestamps t are assigned to each PMID and are used to determine when they became publicly available. We use these timestamps to track how often was a pair of UMLS CUIs \((m_i, m_j)\) appearing in the biomedical literature over time. As a result, we have a list of biologically relevant cross-referenced UMLS CUI co-occurrences, each connected to all PMIDs containing them.
This list is then split into edge lists \(E_t\) , such that each edge list contains pairs \((m_i, m_j)\) added in or before year t . These edge lists are then transformed to dynamic network G with T snapshots:
where \(N_t\) and \(E_t\) represent the set of unique UMLS CUIs (nodes) and their cross-referenced abstract co-occurrences (edges), respectively, and t is the annual timestamp (time resolution can be changed as needed), such that \(G_{t}\) is constructed from all MEDLINE records published before t (e.g., \(t=2011\) ). All networks \(G_{t}\) are simple and undirected.
For each timestamp t , \(G_{t}\) represents a cumulative network, including all the information from \(G_{t-1}\) and new information added in year t .
Tracking the edge importance of time-sliced graphs
We enrich the proposed benchmarking strategy with the information about associations importance at each time step t . In the context of scientific discovery, the importance may be considered from several different perspectives, e.g., as an the influence of an individual finding on future discoveries. In this section we take three different perspectives into account and then combine them together to obtain a final importance score, which we later use to evaluate different hypothesis generation systems with respect to their ability to predict the important associations.
Integrated gradients pipeline
In this step we obtain the information about how edges from graph \(G_t\) influence the appearance of new edges in \(G_{t+1}\) . For that we train a machine learning model, which is able to perform link predictions and then we use an attribution method called Integrated Gradients (IG).
In general, IG is used to understand input features importance with respect to the output a given predictor model produces. In case of link prediction problem, a model outputs likelihood of two nodes \(m_i\) and \(m_j\) being connected for a given network \(G_t\) . The input features for a link prediction model will include the adjacency matrix of \(G_t\) , \(A_t\) , and the predictions themselves can be drawn from a list of edges appearing in the next timestamp \(t + 1\) . If IG is applied to this particular problem, it will provide attribution values for each element of \(A_t\) , which can be reformulated as the importance of edges existing at the timestamp t with respect to their contribution to predicting the edges added at the next timestamp \(t+1\) . This could be interpreted as the influence of current dynamic network structural elements on the information that will be added in future.
Link prediction problem In our setting, the link prediction problem is formulated as following:
We note that predictions of edges \(\hat{E}_{t+1}\) are performed only for nodes \(N_t\) from the graph \(G_t\) at year t .
Adding Node and Edge Features : To enrich the dynamic network G with non-redundant information extracted from text, we add node features and edge weights. Node features are required for Graph Neural Network-based predictor training, which we use in the proposed pipeline.
Node features : Node features are added to each \(G_t\) by applying word2vec algorithm [ 30 ] to the corresponding snapshot of MEDLINE dataset obtained for a timestamp t . In order to perform cleaning and normalization, we replace all tokens in the input texts by their corresponding UMLS CUIs obtained at the NER stage. It significantly reduces the vocabulary size, automatically removing stop-words and enabling vocabulary-guided phrase mining [ 31 ]. It is important to note that each node m has a different vector representation for each time stamp t , which we can refer to as n 2 v ( m , t ).
Edge features (weights) : For simplicity, edge weights are constructed by counting the number of MEDLINE records mentioning a pair of concepts \(e \in E_{t}\) . In other words, for each pair \(e = (m_i, m_j)\) we assign a weight representing the total number of mentions for a pair e in year t .
GNN training
We use a graph neural network-based encoder-decoder architecture. Its encoder consists of two graph convolutional layers [ 32 ] and produces an embedding for each graph node. Decoder takes the obtained node embeddings and outputs the sum of element-wise multiplication of encoded node representations as a characteristic of each pair of nodes.
Attribution
To obtain a connection between newly introduced edges \(\hat{E}_{t+1}\) and existing edges \(E_t\) , we use an attribution method Integrated Gradients (IG) [ 33 ]. It is based on two key assumptions:
Sensitivity: any change in input that affects the output gets a non-zero attribution;
Implementation Invariance: attribution is consistent with the model’s output, regardless of the model’s architecture.
The IG can be applied to a wide variety of ML models as it calculates the attribution scores with respect to input features and not the model weights/activations, which is important, because we focus on relationships between the data points and not the model internal structure.
The integrated gradient (IG) score along \(i^{th}\) dimension for an input x and baseline \(x'\) is defined as:
where \(\frac{\partial F(x)}{\partial x_i}\) is the gradient of F ( x ) along \(i^{th}\) dimension. In our case, input x is the adjacency matrix of \(G_t\) filled with 1 s as default values (we provide all edges \(E_t \in G_t\) ) and baseline \(x'\) is the matrix of zeroes. As a result, we obtain an adjacency matrix \(A(G_t)\) filled with attribution values for each edge \(E_t\) .
Graph-based measures
Betweenness Centrality In order to estimate the structural importance of selected edges, we calculate their betweenness centrality [ 34 ]. This importance measure shows the amount of information passing through the edges, therefore indicating their influence over the information flow in the network. It is defined as
where \(\sigma _{st}\) —the number of shortest paths between nodes s and t ; \(\sigma _{st}(e)\) —the number of shortest paths between nodes s and t passing through edge e .
To calculate the betweenness centrality with respect to the future connections, we restrict the set of vertices V to only those, that are involved in future connections we would like to use for explanation.
Eigenvector Centrality Another graph-based structural importance metric we use is the eigenvector centrality. The intuition behind it is that a node of the network is considered important if it is close to other important nodes. It can be found as a solution of the eigenvalue problem equation:
where A is the network weighted adjacency matrix. Finding the eigenvector corresponding to the largest eigenvalue gives us a list of centrality values \(C_E(v)\) for each vertex \(v \in V\) .
However, we are interested in edge-based metric, which we obtain by taking an absolute difference between the adjacent vertex centralities:
where \(e=(u,v)\) . The last step is to connect this importance measure to time snapshot, which we do by taking a time-base difference between edge-based eigenvector centralities
This metric gives us the eigenvector centrality change with respect to future state of the dynamic graph ( \(t+1\) ).
Second Order Jaccard Similarity One more indicator of how important a particular newly discovered network connection is related to its adjacent nodes neighborhood similarity. The intuition is that more similar their neighborhood is, more trivial the connection is, therefore, it is less important.
We consider a second-order Jaccard similarity index for a given pair of nodes \(m_i\) and \(m_j\) :
Second-order neighborhood of a node u is defined by:
where w iterates over all neighbors of u and N ( w ) returns the neighbors of w .
The second order gives a much better “resolution” or granularity for different connections compared to first-order neighborhood. We also note that it is calculated for a graph \(G_{t-1}\) for all edges \(\hat{E}_{t}\) (before these edges were discovered).
Literature-based measures
Cumulative citation counts Another measure of a connection importance is related to bibliometrics. At each moment in time for each targeted edge we can obtain a list of papers mentioning this edge.
We also have access to a directed citation network, where nodes represent documents and edges represent citations: edges connect one paper to all the papers that it cites. Therefore, the number of citations of a specific paper would equal to in-degree of a corresponding node in a citation network.
To connect paper citations to concepts connections, we compute the sum of citation counts of all papers mentioning a specific connection. Usually, the citation counts follow heavy-tailed distributions (e.g., power law) and counting them at the logarithmic scale is a better practice. However, in our case the citation counts are taken “as-is” to emphasize the difference between the number of citations and the number of mentions. This measure shows the overall citation-based impact of a specific edge over time. The citation information comes from the citation graph, which is consistent with the proposed dynamic network in terms of time slicing methodology.
Combined importance measure for ranking connections
To connect all the components of the importance measure I for edge e , we use the mean percentile rank (PCTRank) of each individual component:
where \(C_i\) is the importance component (one of the described earlier, C —set of all importance components). The importance measure is calculated for each individual edge in graph for each moment in time t with respect to its future (or previous) state \(t+1\) (or \(t-1\) ). Using the mean percentile rank guarantees that the component will stay within a unit interval. The measure I is used to implement an importance-based stratification strategy for benchmarking, as it is discussed in Results section.
In this section we describe the experimental setup and propose a methodology based on different stratification methods. This methodology is unique for the proposed benchmark, because each record is supplied with additional information giving a user more flexible evaluation protocol.
Data collection and processing
Dynamic graph construction.
The numbers of concepts and their associations successfully mapped to UMLS CUI \((m_i, m_j)\) from each dataset are summarized in Table 1 . The number of associations with respect to time is shown in Fig. 2 . It can be seen that the number of concept associations steadily and consistently grows for every subsequent year.
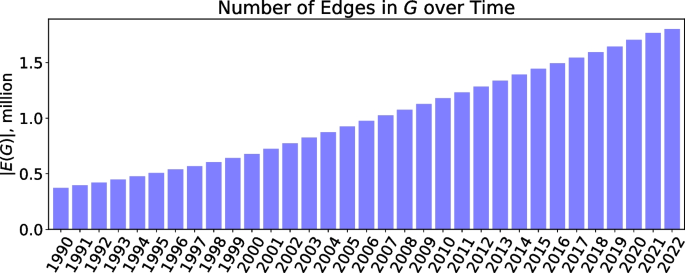
Number of edges in the network G over time. The numbers are reported in millions. Each edge represents a pair of cross-referenced UMLS CUI concepts \((m_i, m_j)\)
Data collection and aggregation is performed in the following pipeline:
All databases are downloaded in their corresponding formats such as comma-separated or Excel spreadsheets, SQL databases or Docker images.
All pairwise interactions in each database are identified.
From all these interactions we create a set of unique concepts, which we then map to UMLS CUIs. Concepts that do not have UMLS representations are dropped.
All original pairwise interactions are mapped with respect to the UMLS codes, as discussed in Databases Processing and Normalization section.
A set of all pairwise interactions is created by merging the mapped interactions from all databases.
This set is then used to find pairwise occurrences in MEDLINE.
Pairwise occurrences found in step 6 are used to construct the main dynamic network G . As it was mentioned earlier, G is undirected and non-attributed (we do not provide types of edges as they are much harder to collect reliably on large scale), which allows us to cover a broader range of pairwise interactions and LBD systems to test. Other pairwise interactions, which are successfully mapped to UMLS CUI, but are not found in the literature, can still be used. They do not have easily identifiable connections to scientific literature and do not contain temporal information, which make them a more difficult target to predict (will be discussed later).
Compound importance calculation
Once the dynamic graph G is constructed, we calculate the importance measure. For that we need to decide on three different timestamps:
Training timestamp: when the predictor models of interest are trained;
Testing timestamp: what moment in time to use to accumulate recently (with respect to step 1) discovered concept associations for models testing;
Importance timestamp: what moment in time to use to calculate the importance measure for concept associations from step 2.
To demonstrate our benchmark, we experiment with different predictive models. In our experimental setup, all models are trained on the data published prior to 2016, tested on associations discovered in 2016 and the importance measure I is calculated based on the most recent fully available timestamp (2022, at the time of writing) with respect to the PubMed annual baseline release. We note that, depending on the evaluation goals, other temporal splits can be used as well. For example, one can decide to evaluate the predictive performance of selected models on more recently discovered connections. For that, they may use the following temporal split: training timestamp—2020, testing timestamp—2021, importance timestamp—2022.
The importance measure I has multiple components, which are described in Methods section. To investigate their relationships and how they are connected to each other, we plot a Spearman correlation matrix showed in Table 2 . Spearman correlation is used because only component’s rank matters in the proposed measure as all components are initially scaled differently.
Evaluation protocol
In our experiments, we demonstrate a scenario for benchmarking hypothesis generation systems. All of the systems are treated as predictors capable of ranking true positive samples (which come from the dynamic network G ) higher than the synthetically generated negatives. The hypothesis generation problem is formulated as binary classification with significant class imbalance.
Evaluation metric
The evaluation metric of choice for our benchmarking is Receiver Operating Characteristic (ROC) curve and its associated Area Under the Curve (AUC), which is calulated as:
where \({\textbf {1}}\) is the indicator function that equals 1 if the score of a negative example \(t_0\) is less than the score of a positive example \(t_1\) ; \(D^0\) , \(D^1\) are the sets of negative and positive examples, respectively. The ROC AUC score quantifies the model’s ability to rank a random positive higher than a random negative.
We note than the scores do not have to be within a specific range, the only requirement is that they can be compared with each other. In fact, using this metric allows us to compare purely classification-based models (such as Node2Vec logistic regression pipeline) and ranking models (like TransE or DistMult), even though the scores of these models may have arbitrary values.
Negative sampling
Our original evaluation protocol can be found in [ 10 ], which is called subdomain recommendation . It is inspired by how biomedical experts perform large-scale experiments to identify the biological instances of interest from a large pool of candidates [ 35 ]. To summarize:
We collect all positive samples after a pre-defined cut date. The data before this cut date is used for prediction system training.
For each positive sample (subject-object pair) we generate N negative pairs, such that the subject is the same and the object in every newly generated pair has the same UMLS semantic type as the object in positive pair;
We evaluate a selected performance measure (ROC AUC) with respect to pairs of semantic types (for example, gene-gene or drug-disease) to better understand domain specific differences.
For this experiment we set \(N=10\) as a trade-off between the evaluation quality and runtime. It can be set higher if more thorough evaluation is needed.
Baseline models description
To demonstrate how the proposed benchmark can be used to evaluate and compare different hypothesis generation system, we use a set of existing models. To make the comparison more fair, all of them are trained on the same snapshots of MEDLINE dataset.
The AGATHA is a general purpose HG system [ 10 , 36 ] incorporates a multi-step pipeline, which processes the entire MEDLINE database of scientific abstracts, constructs a semantic graph from it and trains a predictor model based on transformer encoder architecture. Besides the algorithmic pipeline, the key difference between AGATHA and other link prediction systems is that AGATHA is an end-to-end hypothesis generation framework, where the link prediction is only one of its components.
Node2Vec-based predictor is trained as suggested in the original publication [ 37 ]. We use a network purely constructed with text-mining-based methods.
Knowledge graph embeddings-based models
Knowledge Graph Embeddings (KGE) models are becoming increasingly popular these days, therefore we include them into our comparison. We use Ampligraph [ 38 ] library to train and query a list of KGE models: TransE, HolE, ComplEx and DistMult.
Evaluation with different stratification
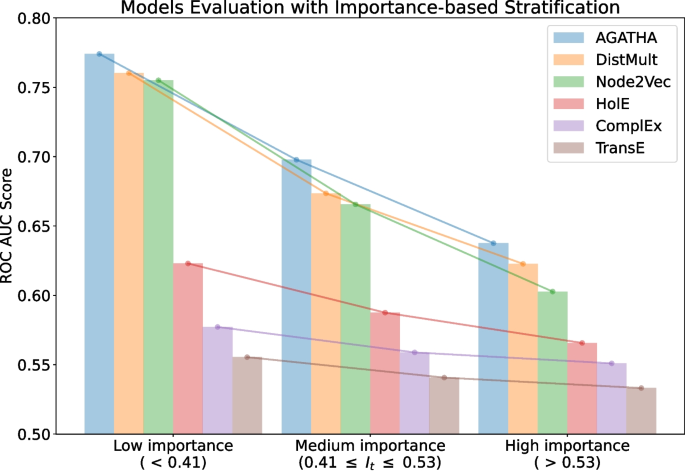
ROC AUC scores for different models trained on the same PubMed snapshot from 2015 and tested on semantic predicates added in 2016 binned with respect to their importance scores
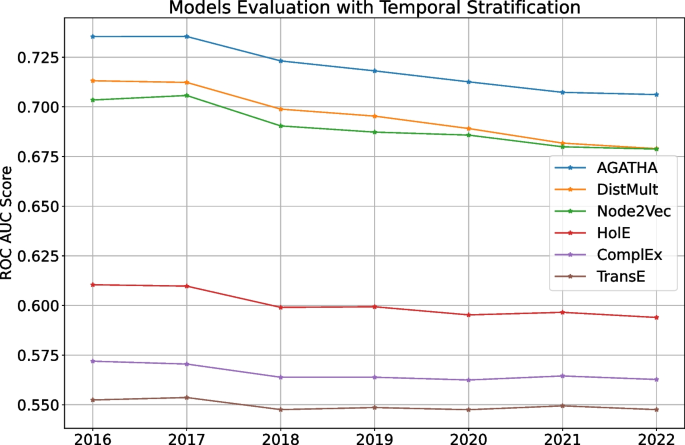
ROC AUC scores for different models trained on the same PubMed snapshot from 2015 and tested on semantic predicates added over time
The proposed benchmarking pipeline enables us to perform different kinds of systems evaluation and comparison with flexibility usually unavailable to other methods. Incorporating both temporal and importance information is helpful to identify trends in models behavior and extend the variety of criteria for domain experts when they decide on a best model suitable for their needs.
Below we present three distinct stratification methods and show how predictor models perform under different evaluation protocols. Even though we use the same performance metric (ROC AUC) across the board, the results differ substantially, suggesting that evaluation strategy plays a significant role in the experimental design.
Semantic stratification
Semantic stratification strategy is the natural way to benchmark hypothesis generation systems, when the goal is to evaluate performance in specific semantic categories. It is especially relevant to the subdomain recommendation problem, which defines our negative sampling procedure. For that we take the testing set of subject-object pairs and group them according to their semantic types and evaluate each group separately (Table 3 ).
Importance-based stratification
The next strategy is based on the proposed importance measure I . This measure ranks all the positive subject-object pairs from the test set and, therefore, can be used to split them into equally-sized bins, according to their importance score. In our experiment, we split the records into three bins, representing low, medium and high importance values. Negative samples are split accordingly. Then each group is evaluated separately. The results of this evaluation are presented in Fig. 3 .
The results indicate that the importance score I could also reflect the difficulty of making a prediction. Specifically, pairs that receive higher importance scores tend to be more challenging for the systems to be identified correctly. In models that generally exhibit high performance (e.g., DistMult), the gap in ROC AUC scores between pairs with low importance scores and those with high importance scores is especially pronounced. The best model in this list is AGATHA as it utilizes the most nuanced hypothesis representation, namely, its transformer architecture is trained to leverage not only node embeddings but also to account for the non-overlapping neighborhoods of concepts.
Temporal stratification
The last strategy shows how different models trained once perform over time . For that we fix the training timestamp on 2015 and evaluate each models on testing timestamps from 2016 to 2022. For clarity, we do not use importance values for this experiment and only focus on how the models perform over time on average . The results are shown in Fig. 4 .
Figure 4 highlights how predictive performance gradually decays over time for every model in the list. This behavior can be expected: the gap between training and testing data increases over time, which makes it more difficult for models to perform well as time goes by. Therefore, it is a good idea to keep the predictor models up-to-date, which we additionally discuss in the next section.
We divide the discussion into separate parts: topics related to evaluation challenges and topics related to different predictor model features. We also describe the challenges and scope for the future work at the end of the section.
Evaluation-based topics
Data collection and processing challenges.
The main challenge of this work comes from the diverse nature of biomedical data. This data may be described in many different ways and natural language may not be the most commonly used. Our results indicate that a very significant part of biocurated connections “flies under the radar” of text-mining systems and pipelines due to several reasons:
Imperfections of text-mining methods;
Multiple standards to describe biomedical concepts;
The diversity of scientific language: many biomedical associations (e.g. gene-gene interactions may be primarily described in terms of co-expression);
Abstracts are not enough for text mining [ 39 ].
The proposed methodology for the most part takes the lowest common denominator approach: we discard concepts not having UMLS representations and associations not appearing in PubMed abstracts. However, our approach still allows us to extract a significant number of concept associations and to use them for quantitative analysis. We should also admit that the aforementioned phenomenon of biomedical data discrepancy leads us to some interesting results, which we discuss below.
Different nature of biomedical DBs and literature-extracted data
The experiment clearly indicates significant differences between different kinds of associations with respect their corresponding data sources in models performance comparison. For this experiment we take one of the evaluated earlier systems (AGATHA 2015) and run the semantically-stratified version of benchmark collected from three different data sources:
Proposed benchmark dataset: concept associations extracted from biocurated databases with cross-referenced literature data;
Concept associations extracted from biocurated databases, but which we could not cross-reference with literature data;
Dataset composed of associations extracted with a text mining framework (SemRep).
Datasets (1) and (3) were constructed from associations found in MEDLINE snapshot from 2020. For dataset (2) it was impossible to identify the time connections were added, therefore the cut date approach was not used. All three datasets were downsampled with respect to the proposed benchmark (1), such that the number of associations is the same across all of them.
The results of this experiment are shown in Table 4 . It is evident that associations extracted from biocurated databases (1) and (2) propose a more significant challenge for a text-mining-based system. Cross-referencing from literature makes sure that similar associations can be at least discovered by these systems at the training time, therefore, the AGATHA performance on dataset (1) is higher compared to dataset (2). These results may indicate that biocurated associations, which cannot be cross-referenced, belong to a different data distribution, and, therefore, purely text mining-based systems fall short due to the limitations of the underlying information extraction algorithms.
Models-related topics
Text mining data characteristics.
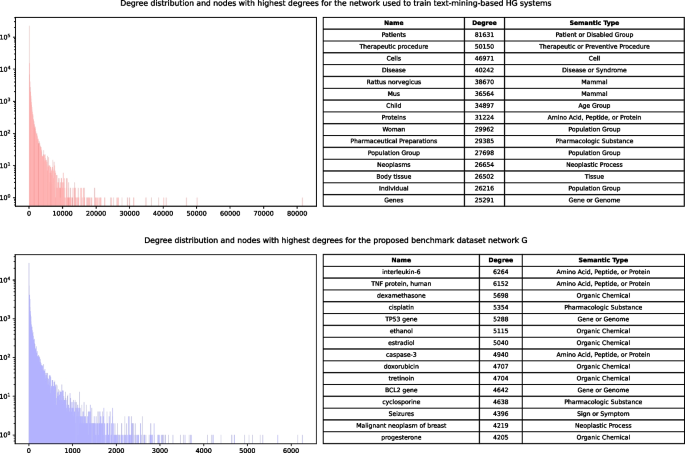
Degree distributions and nodes with highest degrees for two networks: the one used for training of text-mining-based predictor models (red, top) and the network G from the proposed benchmark dataset (blue, bottom)
In order to demonstrate the differences between biologically curated and text mining-based knowledge, we can consider their network representations.
The network-based models we show in this work are trained on text-mining-based networks, which are built on top of semantic predicates extracted from a NLP tool SemRep. This tool takes biomedical text as input and extracts triples (subject-verb-object) from the text and performs a number of additional tasks, such as:
Named Entity Recognition
Concept Normalization
Co-reference Resolution
and some others. This tool operates on UMLS Metathesaurus, one of the largest and most diverse biomedical thesaurus, including many different vocabularies.
The main problem of text-mining tools like SemRep is that they tend to produce noisy (and often not quite meaningful from the biomedical prospective) data. As a result, the underlying data that is used to build and validate literature-based discovery systems may not represent the results that domain experts expect to see.
However, these systems are automated and, therefore, are widely used as a tool to extract information from literature in uninterrupted manner. Then this information is used for training different kinds of predictors (either rule-based, statistical or deep learning).
To demonstrate this phenomenon, we compare two networks, where nodes are biomedical terms and edges are associations between them. The difference between them lies in their original data source, which is either:
PubMed abstracts processed with SemRep tool;
Biocurated databases, which connections are mapped to pairs of UMLS CUI terms and cross-referenced with MEDLINE records.
Connections from the network (2) are used in the main proposed benchmarking framework (network G ). The comparison is shown in Fig. 5 as a degree distribution of both networks. We can see that network (1) has a small number of very high-degree nodes. These nodes may affect negatively to the overall predictive power of any model using networks like (1) as a training set, because they introduce a large number of “shortcuts” to the network, which do not have any significant biological value. We also show the top most high-degree nodes for both networks. For the network (1), all of them appear to be very general and most of them (e.g. “Patients” or “Pharmaceutical Preparations”) can be described as noise. Network (2), in comparison, contain real biomedical entities, which carry domain-specific meaning.
Training data threshold influence
As the Temporal Stratification experiment in the Results section suggests, the gap between training and testing timestamps plays a noticeable role in models predictive performance.
To demonstrate this phenomena from a different perspective, we now fix the testing timestamp and vary the training timestamp. We use two identical AGATHA instances, but trained on different MEDLINE snapshots: 2015 and 2020. The testing timestamp for this experiment is 2021, such that none of the models has access to the test data.
The results shown in Table 5 illustrate that having more recent training data does not significantly increase model’s predictive power for the proposed benchmark. This result may be surprising, but there is a possible explanation: a model learns the patterns from the training data distribution and that data distribution stays consistent for both training cut dates (2015 and 2020). However, that does not mean that the data distribution in the benchmark behaves the same way. In fact, it changes with respect to both data sources: textual and DB-related.
Semantic types role in predictive performance
Another aspect affecting models predictive performance is having access to domain information. Since we formulate the problem as subdomain recommendation, knowing concept-domain relationships may be particularly valuable. We test this idea by injecting semantic types information into the edge type for tested earlier Knowledge Graph Embedding models. As opposed to classic link prediction methods (such as node2vec), Knowledge Graph modeling was designed around typed edges and allows this extension naturally.
Results in Table 6 show that semantic type information provides a very significant improvement for models predictive performance.
Large language models for scientific discovery
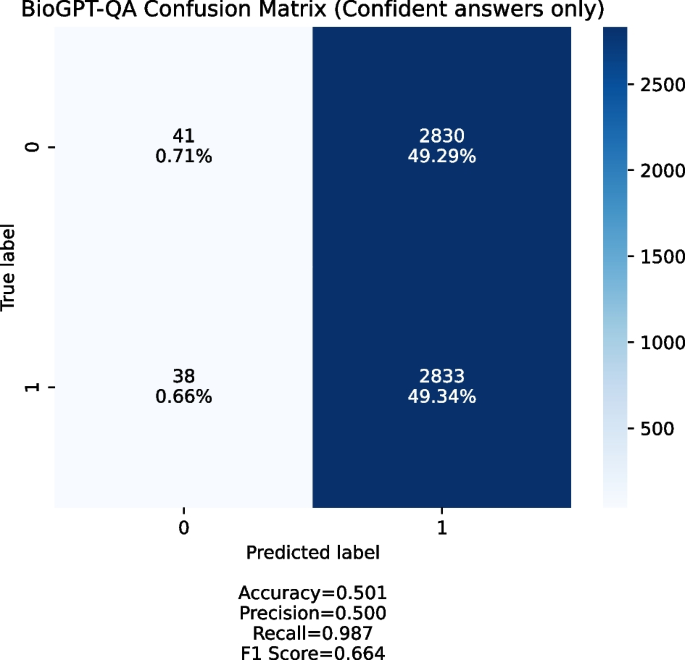
Confusion matrix obtained by the BioGPT-QA model. Only confident answers (Yes/No) were taken into account
Recent advances in language model development raised a logical question about usefulness of these models in scientific discovery, especially in biomedical area [ 40 ]. Problems like drug discovery, drug repurposing, clinical trial optimization and many others may benefit significantly from systems trained on a large amount of scientific biomedical data.
Therefore, we decide to test how one of these systems would perform in our benchmark. We take one of the recently released generative pre-trained transformer models BioGPT [ 4 ] and run a set of test queries.
BioGPT model was chosen due to the following reasons:
It is recently released (2022);
It includes fine-tuned models, which show good performance on downstream tasks;
It is open source and easily accessible.
We use a BioGPT-QA model to perform the benchmarking, because it was fine-tuned on PubMedQA [ 41 ] dataset and outputs the answer as yes/maybe/no, which is easy to parse and represent as a (binary) classifier output.
The question prompt was formulated as the following: “Is there a relationship between <term 1> and <term 2>?”. PubMedQA format also requires a context from a PubMed abstract, which does not exist in our case, because it is a discovery problem. However, we supply an abstract-like context, which is constructed by concatenating term definitions extracted from UMLS Metathesaurus for both source and target terms.
A sample prompt looks like this: “Is there a relationship between F1-ATPase and pyridoxal phosphate? context: F1-ATPase—The catalytic sector of proton-translocating ATPase complexes. It contains five subunits named alpha, beta, gamma, delta and eta. pyridoxal phosphate—This is the active form of VITAMIN B6 serving as a coenzyme for synthesis of amino acids, neurotransmitters (serotonin, norepinephrine), sphingolipids, aminolevulinic acid...”
When we ran the experiment, we noticed two things:
BioGPT is often not confident in its responses, which means that it outputs “maybe” or two answers (both “yes” and “no”) for about 40% of the provided queries;
The overwhelming majority of provided queries are answered positively when the answer is confident.
Figure 6 shows a confusion matrix for queries with confident answer. We generate the queries set with 1:1 positive to negative ratio. Most of the answers BioGPT-QA provides are positive, which means that the system produces too many false positives and is not usable in the discovery setting.
Challenges in benchmarking for hypothesis generation
Binary interactions. Not every discovery can be represented as a pair of terms, but this is something that most of biomedical graph-based knowledge discovery systems work with. It is a significant limitation of the current approach and a motif discovery is a valid potential direction for future work. Moreover, many databases represent their records as binary interactions [ 42 , 43 , 44 , 45 , 46 ], which can be easily integrated into a link prediction problem.
Directionality. Currently, our choice for pairwise interactions is to omit the directionality information to allow more systems to be evaluated with our framework and cover more pairwise interactions. Directionality is an important component of pairwise interactions, especially when they have types and are formulated in a predication form as a triple: (subject-predicate-object) . Currently, we omit the predicate part and only keep pairs of terms for easier generalization. In many cases, a uni-directional edge \(i\rightarrow j\) does not imply non-existence of \(i\leftarrow j\) . Moreover, in the low-dimensional graph representation construction it is clearly preferable to use undirected edges in our context due to the scarcity of biomedical information. Another caveat is that the tools that detect the logical direction of the predicate in the texts are not perfect [ 47 ]. The information about each particular direction can still be recovered from the underlying cross-referencing citations.
Concept normalization . UMLS is a powerful system combining many biomedical vocabularies together. However, it has certain limitations, such as relatively small number of proteins and chemical compounds. We also observe that many UMLS terms are never covered in the scientific abstracts, even though they exist in the Metathesaurus. This limits the number of obtainable interactions significantly. However, UMLS covers many areas of biomedicine, such as genes, diseases, proteins, chemicals and many others and also provides rich metadata. In addition, NLM provides software for information extraction. There are other vocabularies, which have greater coverage in certain areas (e.g., UniProt ID for proteins or PubChem ID for chemicals), but their seamless integration into a heterogeneous network with literature poses additional challenges that will be gradually addressed in the future work.
We have developed and implemented a comprehensive benchmarking system Dyport for evaluating biomedical hypothesis generation systems. This benchmarking system is advancing the field by providing a structured and systematic approach to assess the efficacy of various hypothesis generation methodologies.
In our pipeline we utilized several curated datasets, which provide a basis in testing the hypothesis generation systems under realistic conditions. The informative discoveries have been integrated into the dynamic graph on top of which we introduced the quantification of discovery importance. This approach allowed us to add a new dimension to the benchmarking process, enabling us to not only assess the accuracy of the hypotheses generated but also their relevance and potential impact in the field of biomedical research. This quantification of discovery importance is a critical step forward, as it aligns the benchmarking process more closely with the practical and applied goals of biomedical research.
We have demonstrated the use case of several graph-based link prediction systems’ verification and concluded that such testing is way more productive than traditional link prediction benchmarks. However, the utility of our benchmarking system extends beyond these examples. We advocate for its widespread adoption to validate the quality of hypothesis generation, aiming to broaden the range of scientific discoveries accessible to the wider research community. Our system is designed to be inclusive, welcoming the addition of more diverse cases.
Future work includes integration of the benchmarking process in the hypothesis system visualization [ 48 ], spreading to other than biomedical areas [ 49 ], integration of novel importance measures, and healthcare benchmarking cases.
Swanson DR. Undiscovered public knowledge. Libr Q. 1986;56(2):103–18.
Article Google Scholar
Swanson DR, Smalheiser NR, Torvik VI. Ranking indirect connections in literature-based discovery: the role of medical subject headings. J Am Soc Inform Sci Technol. 2006;57(11):1427–39.
Article CAS Google Scholar
Peng Y, Bonifield G, Smalheiser N. Gaps within the biomedical literature: Initial characterization and assessment of strategies for discovery. Front Res Metrics Anal. 2017;2:3.
Luo R, Sun L, Xia Y, Qin T, Zhang S, Poon H, Liu T-Y. Biogpt: generative pre-trained transformer for biomedical text generation and mining. Brief Bioinform. 2022;23(6):409.
Sybrandt J, Safro I. Cbag: conditional biomedical abstract generation. PLoS ONE. 2021;16(7):0253905.
Sybrandt J, Shtutman M, Safro I. Moliere: automatic biomedical hypothesis generation system. In: Proceedings of the 23rd ACM SIGKDD. KDD ’17, 2017. pp. 1633–1642. ACM, New York, NY, USA. https://doi.org/10.1145/3097983.3098057 .
Sedler AR, Mitchell CS. Semnet: using local features to navigate the biomedical concept graph. Front Bioeng Biotechnol. 2019;7:156.
Article PubMed PubMed Central Google Scholar
Hristovski D, Peterlin B, Mitchell JA, Humphrey SM. Using literature-based discovery to identify disease candidate genes. Int J Med Inform. 2005;74(2):289–98.
Article PubMed Google Scholar
Gordon MD, Dumais S. Using latent semantic indexing for literature based discovery. J Am Soc Inf Sci. 1998;49(8):674–85.
Sybrandt J, Tyagin I, Shtutman M, Safro I. AGATHA: automatic graph mining and transformer based hypothesis generation approach. In: Proceedings of the 29th ACM international conference on information and knowledge management, 2020;2757–64.
Sourati J, Evans J. Accelerating science with human-aware artificial intelligence. Nat Hum Behav. 2023;7:1682–96.
Chen Y, Argentinis JE, Weber G. IBM Watson: how cognitive computing can be applied to big data challenges in life sciences research. Clin Ther. 2016;38(4):688–701.
Xun G, Jha K, Gopalakrishnan V, Li Y, Zhang A. Generating medical hypotheses based on evolutionary medical concepts. In: 2017 IEEE International conference on data mining (ICDM), pp. 535–44 (2017). https://doi.org/10.1109/ICDM.2017.63 .
Cameron D, Kavuluru R, Rindflesch TC, Sheth AP, Thirunarayan K, Bodenreider O. Context-driven automatic subgraph creation for literature-based discovery. J Biomed Inform. 2015;54:141–57. https://doi.org/10.1016/j.jbi.2015.01.014 .
Sebastian Y, Siew E-G, Orimaye SO. Learning the heterogeneous bibliographic information network for literature-based discovery. Knowl-Based Syst. 2017;115:66–79.
Miranda A, Mehryary F, Luoma J, Pyysalo S, Valencia A, Krallinger M. Overview of drugprot biocreative vii track: quality evaluation and large scale text mining of drug-gene/protein relations. In: Proceedings of the seventh biocreative challenge evaluation workshop, 2021;11–21.
Breit A, Ott S, Agibetov A, Samwald M. OpenBioLink: a benchmarking framework for large-scale biomedical link prediction. Bioinformatics. 2020;36(13):4097–8. https://doi.org/10.1093/bioinformatics/btaa274 .
Article CAS PubMed Google Scholar
Sybrandt J, Shtutman M, Safro I. Large-scale validation of hypothesis generation systems via candidate ranking. In: 2018 IEEE international conference on big data, 2018; 1494–1503. https://doi.org/10.1109/bigdata.2018.8622637 .
Kilicoglu H, Shin D, Fiszman M, Rosemblat G, Rindflesch TC. Semmeddb: a pubmed-scale repository of biomedical semantic predications. Bioinformatics. 2012;28(23):3158–60.
Article CAS PubMed PubMed Central Google Scholar
Fannjiang C, Listgarten J. Is novelty predictable? Cold Spring Harb Perspect Biol. 2024;16: a041469.
Jeon D, Lee J, Ahn J, Lee C. Measuring the novelty of scientific publications: a fastText and local outlier factor approach. J Inform. 2023;17: 101450.
Small H, Tseng H, Patek M. Discovering discoveries: Identifying biomedical discoveries using citation contexts. J Inform. 2017;11:46–62.
Bordes A, Usunier N, Garcia-Duran A, Weston J, Yakhnenko O. Translating embeddings for modeling multi-relational data. In: Advances in neural information processing systems, 2013; 2787–2795.
Bonner S, Barrett IP, Ye C, Swiers R, Engkvist O, Hoyt CT, Hamilton WL. Understanding the performance of knowledge graph embeddings in drug discovery. Artif Intell Life Sci. 2022;2: 100036.
Google Scholar
Bonner S, Barrett IP, Ye C, Swiers R, Engkvist O, Bender A, Hoyt CT, Hamilton WL. A review of biomedical datasets relating to drug discovery: a knowledge graph perspective. Brief Bioinform. 2022;23(6):404.
Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2015;44(D1):457–62. https://doi.org/10.1093/nar/gkv1070 .
Bodenreider O. The unified medical language system (umls): integrating biomedical terminology. Nucleic Acids Res. 2004;32(suppl_1):267–70.
Rindflesch TC, Fiszman M. The interaction of domain knowledge and linguistic structure in natural language processing: interpreting hypernymic propositions in biomedical text. J Biomed Inform. 2003;36(6):462–77. https://doi.org/10.1016/j.jbi.2003.11.003 .
Xing R, Luo J, Song T. Biorel: towards large-scale biomedical relation extraction. BMC Bioinform. 2020;21(16):1–13.
Mikolov T, Sutskever I, Chen K, Corrado GS, Dean J. Distributed representations of words and phrases and their compositionality. Adv Neural Inf Process Syst 2013;26.
Aronson AR. Effective mapping of biomedical text to the umls metathesaurus: the metamap program. In: Proceedings of the AMIA symposium, 2001;p. 17.
Welling M, Kipf TN. Semi-supervised classification with graph convolutional networks. In: Journal of international conference on learning representations (ICLR 2017), 2016.
Sundararajan M, Taly A, Yan Q. Axiomatic attribution for deep networks. In: International conference on machine learning, pp. 3319–3328, 2017.
Brandes U. A faster algorithm for betweenness centrality. J Math Sociol. 2001;25(2):163–77.
Aksenova M, Sybrandt J, Cui B, Sikirzhytski V, Ji H, Odhiambo D, Lucius MD, Turner JR, Broude E, Peña E, et al. Inhibition of the dead box rna helicase 3 prevents hiv-1 tat and cocaine-induced neurotoxicity by targeting microglia activation. J Neuroimmune Pharmacol. 2019;1–15.
Tyagin I, Kulshrestha A, Sybrandt J, Matta K, Shtutman M, Safro I. Accelerating covid-19 research with graph mining and transformer-based learning. In: Proceedings of the AAAI conference on artificial intelligence, 2022;36:12673–9.
Grover A, Leskovec J. Node2vec: Scalable feature learning for networks. In: Proceedings of the 22nd ACM SIGKDD international conference on knowledge discovery and data mining. KDD ’16, 2016, pp. 855–864. Association for Computing Machinery, New York. https://doi.org/10.1145/2939672.2939754 .
Costabello L, Bernardi A, Janik A, Pai S, Van CL, McGrath R, McCarthy N, Tabacof P. AmpliGraph: a library for representation learning on knowledge graphs, 2019. https://doi.org/10.5281/zenodo.2595043 .
Sybrandt J, Carrabba A, Herzog A, Safro I. Are abstracts enough for hypothesis generation? In: 2018 IEEE international conference on big data, 2018;1504–1513. https://doi.org/10.1109/bigdata.2018.8621974 .
Liu Z, Roberts RA, Lal-Nag M, Chen X, Huang R, Tong W. Ai-based language models powering drug discovery and development. Drug Discovery Today. 2021;26(11):2593–607.
Jin Q, Dhingra B, Liu Z, Cohen WW, Lu X. Pubmedqa: a dataset for biomedical research question answering, 2019; arXiv preprint arXiv:1909.06146 .
Davis AP, Wiegers TC, Johnson RJ, Sciaky D, Wiegers J, Mattingly CJ. Comparative toxicogenomics database (ctd): update 2023. Nucleic Acids Res. 2022. https://doi.org/10.1093/nar/gkac833 .
Piñero J, Ramírez-Anguita JM, Saüch-Pitarch J, Ronzano F, Centeno E, Sanz F. Furlong LI The DisGeNET knowledge platform for disease genomics: 2019 update. Nucleic Acids Res. 2019;48(D1):845–55. https://doi.org/10.1093/nar/gkz1021 .
Ursu O, Holmes J, Knockel J, Bologa CG, Yang JJ, Mathias SL, Nelson SJ, Oprea TI. DrugCentral: online drug compendium. Nucleic Acids Research. 2016;45(D1):932–9. https://doi.org/10.1093/nar/gkw993 .
Calderone A, Castagnoli L, Cesareni G. Mentha: a resource for browsing integrated protein-interaction networks. Nat Methods. 2013;10(8):690–1.
Zeng K, Bodenreider O, Kilbourne J, Nelson SJ. Rxnav: a web service for standard drug information. In: AMIA annual symposium proceedings, 2006; vol. 2006, p. 1156.
Kilicoglu H, Rosemblat G, Fiszman M, Shin D. Broad-coverage biomedical relation extraction with SemRep. BMC Bioinform. 2020;21:1–28.
Tyagin I, Safro I. Interpretable visualization of scientific hypotheses in literature-based discovery. BioCretive Workshop VII; 2021. https://www.biorxiv.org/content/10.1101/2021.10.29.466471v1 .
Marasco D, Tyagin I, Sybrandt J, Spencer JH, Safro I. Literature-based discovery for landscape planning, 2023. arXiv preprint arXiv:2306.02588 .
Rehurek R, Sojka P. Gensim-python framework for vector space modelling. NLP Centre, Faculty of Informatics, Masaryk University, Brno, Czech Republic 2011;3(2).
Fey M, Lenssen JE. Fast graph representation learning with PyTorch Geometric. In: ICLR workshop on representation learning on graphs and manifolds, 2019.
Kokhlikyan, N., Miglani, V., Martin, M., Wang, E., Alsallakh, B., Reynolds, J., Melnikov, A., Kliushkina, N., Araya, C., Yan, S., Reblitz-Richardson, O. Captum: a unified and generic model interpretability library for PyTorch, 2020.
Sollis E, Mosaku A, Abid A, Buniello A, Cerezo M, Gil L, Groza T, Güneş O, Hall P, Hayhurst J, Ibrahim A, Ji Y, John S, Lewis E, MacArthur JL, McMahon A, Osumi-Sutherland D, Panoutsopoulou K, Pendlington Z, Ramachandran S, Stefancsik R, Stewart J, Whetzel P, Wilson R, Hindorff L, Cunningham F, Lambert S, Inouye M, Parkinson H, Harris L. The NHGRI-EBI GWAS catalog: knowledgebase and deposition resource. Nucleic Acids Res. 2022;51(D1):977–85. https://doi.org/10.1093/nar/gkac1010 .
Szklarczyk D, Gable AL, Nastou KC, Lyon D, Kirsch R, Pyysalo S, Doncheva NT, Legeay M, Fang T, Bork P, Jensen LJ, von Mering C. The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Research. 2020;49(D1):605–12. https://doi.org/10.1093/nar/gkaa1074 .
Fricke S. Semantic scholar. J Med Lib Assoc: JMLA. 2018;106(1):145.
Download references
Acknowledgements
We would like to thank two anonymous referees whose thoughtful comments helped to improve the paper significantly. This research was supported by NIH award #R01DA054992. The computational experiments were supported in part through the use of DARWIN computing system: DARWIN—A Resource for Computational and Data-intensive Research at the University of Delaware and in the Delaware Region, which is supported by NSF Grant #1919839.
This research was supported by NIH award #R01DA054992. The computational experiments were supported in part through the use of DARWIN computing system: DARWIN—A Resource for Computational and Data-intensive Research at the University of Delaware and in the Delaware Region, which is supported by NSF Grant #1919839.
Author information
Authors and affiliations.
Center for Bioinformatics and Computational Biology, University of Delaware, Newark, DE, 19713, USA
Ilya Tyagin
Department of Computer and Information Sciences, University of Delaware, Newark, DE, 19716, USA
You can also search for this author in PubMed Google Scholar
Contributions
IT processed and analyzed the textual and database data, trained models and implemented the computational pipeline. IS formulated the main idea, supervised the project and provided feedback. Both authors contributed to writing, read and approved the final manuscript.
Corresponding authors
Correspondence to Ilya Tyagin or Ilya Safro .
Ethics declarations
Competing interests.
I declare that the authors have no Conflict of interest as defined by BMC, or other interests that might be perceived to influence the results and/or discussion reported in this paper.
Availability of data and materials
The dataset(s), materials and code supporting the conclusions of this article is(are) available in the GitHub repository: https://github.com/IlyaTyagin/Dyport .
Additional information
Publisher's note.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Appendix A: Incorporated technologies
To construct the benchmark, we propose a multi-step pipeline, which requires several key technologies to be used. For the text mining part, we use SemRep [ 28 ] and gensim [ 50 ] implementation of word2vec algorithm. For further stages involving graph learning, we utilize Pytorch Geometric framework and Captum explainability library.
UMLS (Unified Medical Language System) [ 27 ] is one of the fundamental technologies provided by NLM, which consolidates and disseminates essential terminology, taxonomies, and coding norms, along with related materials, such as definitions and semantic types. UMLS is used in the proposed work as a system of concept unique identifiers (CUI) bringing together terms from different vocabularies.
SemRep [ 47 ] is an NLM-developed software, performing extraction of semantic predicates from biomedical texts. It also has the named entity recognition (NER) capabilities (based on MetaMap [ 31 ] backend) and automatically performs entity normalization based on the context.
Word2Vec [ 30 ] is an approach for creating efficient word embeddings. It was proposed in 2013 and is proven to be an excellent technique for generating static (context-independent) latent word representations. The implementation used in this work is based on gensim [ 50 ] library.
Pytorch geometric (PyG) [ 51 ] library is built on top of Pytorch framework focusing on graph geometric learning. It implements a variety of algorithms from published research papers, supports arbitrary-scaled graphs and is well integrated into Pytorch ecosystem. We use PyG to train a graph neural network (GNN) for link prediction problem, which we explain in more detail in methods section.
Captum [ 52 ] package is an extension of Pytorch enabling the explainability of many ML models. It contains attribution methods, such as saliency maps, integrated gradients, Shapley value sampling and others. Captum is supported by PyG library and used in this work to calculate attributions of the proposed GNN.
Appendix B: Incorporated data sources
We review and include a variety of biomedical databases, containing curated connections between different kinds of entities.
KEGG (Kyoto Encyclopedia of Genes and Genomes) [ 26 ] is a collection of resources for understanding principles of work of biological systems (such as cells, organisms or ecosystems) and offering a wide variety of entry points. One of the main components of KEGG is a set of pathway maps, representing molecular interactions as network diagrams.
CTD (The Comparative Toxicogenomics Database) [ 42 ] is a publicly available database focused on collecting the information about environmental exposures effects on human health.
DisGenNET [ 43 ] is a discovery platform covering genes and variants and their connections to human diseases. It integrates data from a list of publicly available databases and repositories and scientific literature.
GWAS (Genome-Wide Association Studies) [ 53 ] is a catalog of human genome-wide association studies, developed by EMBL-EBI and NHGRI. Its aim is to identify and systematize associations of genotypes with phenotypes across human genome.
STRING [ 54 ] is a database aiming to integrate known and predicted protein associations, both physical and functional. It utilizes a network-centric approach and assigns a confidence score for all interactions in the network based on the evidence coming from different sources: text mining, computational predictions and biocurated databases.
DrugCentral [ 44 ] is an online drug information resource aggregating information about active ingredients, indications, pharmacologic action and other related data with respect to FDA, EMA and PMDA-approved drugs.
Mentha [ 45 ] is an evidence-based protein interaction browser (and corresponding database), which takes advantage of International Molecular Exchange (IMEx) consortium. The interactions are curated by experts in compliance with IMEx policies enabling regular weekly updates. Compared to STRING, Mentha is focused on precision over comprehensiveness and excludes any computationally predicted records.
RxNav [ 46 ] is a web-service providing an integrated view on drug information. It contains the information from NLM drug terminology RxNorm, drug classes RxClass and drug-drug interactions collected from ONCHigh and DrugBank sources.
Semantic scholar [ 55 ] is a search engine and research tool for scientific papers developed by the Allen Institute for Artificial Intelligence (AI2). It provides rich metadata about publications which enables us to use Semantic Scholar data for network-based citation analysis.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/ . The Creative Commons Public Domain Dedication waiver ( http://creativecommons.org/publicdomain/zero/1.0/ ) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
Reprints and permissions
About this article
Cite this article.
Tyagin, I., Safro, I. Dyport: dynamic importance-based biomedical hypothesis generation benchmarking technique. BMC Bioinformatics 25 , 213 (2024). https://doi.org/10.1186/s12859-024-05812-8
Download citation
Received : 31 January 2024
Accepted : 16 May 2024
Published : 13 June 2024
DOI : https://doi.org/10.1186/s12859-024-05812-8
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Hypothesis Generation
- Literature-based Discovery
- Link Prediction
- Benchmarking
- Natural Language Processing
BMC Bioinformatics
ISSN: 1471-2105
- General enquiries: [email protected]

Scientific hypothesis generation process in clinical research: a secondary data analytic tool versus experience study protocol
- Find this author on Google Scholar
- Find this author on PubMed
- Search for this author on this site
- ORCID record for Xia Jing
- For correspondence: [email protected]
- Info/History
- Supplementary material
- Preview PDF
Background Scientific hypothesis generation is a critical step in scientific research that determines the direction and impact of any investigation. Despite its vital role, we have limited knowledge of the process itself, hindering our ability to address some critical questions.
Objective To what extent can secondary data analytic tools facilitate scientific hypothesis generation during clinical research? Are the processes similar in developing clinical diagnoses during clinical practice and developing scientific hypotheses for clinical research projects? We explore the process of scientific hypothesis generation in the context of clinical research. The study is designed to compare the role of VIADS, our web-based interactive secondary data analysis tool, and the experience levels of study participants during their scientific hypothesis generation processes.
Methods Inexperienced and experienced clinical researchers are recruited. In this 2×2 study design, all participants use the same data sets during scientific hypothesis-generation sessions, following pre-determined scripts. The inexperienced and experienced clinical researchers are randomly assigned into groups with and without using VIADS. The study sessions, screen activities, and audio recordings of participants are captured. Participants use the think-aloud protocol during the study sessions. After each study session, every participant is given a follow-up survey, with participants using VIADS completing an additional modified System Usability Scale (SUS) survey. A panel of clinical research experts will assess the scientific hypotheses generated based on pre-developed metrics. All data will be anonymized, transcribed, aggregated, and analyzed.
Results This study is currently underway. Recruitment is ongoing via a brief online survey 1 . The preliminary results show that study participants can generate a few to over a dozen scientific hypotheses during a 2-hour study session, regardless of whether they use VIADS or other analytic tools. A metric to assess scientific hypotheses within a clinical research context more accurately, comprehensively, and consistently has also been developed.
Conclusion The scientific hypothesis-generation process is an advanced cognitive activity and a complex process. Clinical researchers can quickly generate initial scientific hypotheses based on data sets and prior experience based on our current results. However, refining these scientific hypotheses is much more time-consuming. To uncover the fundamental mechanisms of generating scientific hypotheses, we need breakthroughs that capture thinking processes more precisely.
Competing Interest Statement
The authors have declared no competing interest.
Clinical Trial
This study is not a clinical trial per NIH definition.
Funding Statement
The project is supported by a grant from the National Library of Medicine of the United States National Institutes of Health (R15LM012941) and partially supported by the National Institute of General Medical Sciences of the National Institutes of Health (P20 GM121342). The content is solely the author's responsibility and does not necessarily represent the official views of the National Institutes of Health.
Author Declarations
I confirm all relevant ethical guidelines have been followed, and any necessary IRB and/or ethics committee approvals have been obtained.
The details of the IRB/oversight body that provided approval or exemption for the research described are given below:
The study has been approved by the Institutional Review Board (IRB) at Clemson University (IRB2020-056).
I confirm that all necessary patient/participant consent has been obtained and the appropriate institutional forms have been archived, and that any patient/participant/sample identifiers included were not known to anyone (e.g., hospital staff, patients or participants themselves) outside the research group so cannot be used to identify individuals.
I understand that all clinical trials and any other prospective interventional studies must be registered with an ICMJE-approved registry, such as ClinicalTrials.gov. I confirm that any such study reported in the manuscript has been registered and the trial registration ID is provided (note: if posting a prospective study registered retrospectively, please provide a statement in the trial ID field explaining why the study was not registered in advance).
I have followed all appropriate research reporting guidelines and uploaded the relevant EQUATOR Network research reporting checklist(s) and other pertinent material as supplementary files, if applicable.
Data Availability
This manuscript is the study protocol. After we analyze and publish the results, transcribed, aggregated, de-identified data can be requested from the authors.
Abbreviations
View the discussion thread.
Supplementary Material
Thank you for your interest in spreading the word about medRxiv.
NOTE: Your email address is requested solely to identify you as the sender of this article.

Citation Manager Formats
- EndNote (tagged)
- EndNote 8 (xml)
- RefWorks Tagged
- Ref Manager
- Tweet Widget
- Facebook Like
- Google Plus One
Subject Area
- Health Informatics
- Addiction Medicine (336)
- Allergy and Immunology (660)
- Anesthesia (177)
- Cardiovascular Medicine (2578)
- Dentistry and Oral Medicine (311)
- Dermatology (218)
- Emergency Medicine (390)
- Endocrinology (including Diabetes Mellitus and Metabolic Disease) (917)
- Epidemiology (12103)
- Forensic Medicine (10)
- Gastroenterology (749)
- Genetic and Genomic Medicine (4008)
- Geriatric Medicine (377)
- Health Economics (670)
- Health Informatics (2588)
- Health Policy (993)
- Health Systems and Quality Improvement (964)
- Hematology (357)
- HIV/AIDS (827)
- Infectious Diseases (except HIV/AIDS) (13588)
- Intensive Care and Critical Care Medicine (785)
- Medical Education (397)
- Medical Ethics (107)
- Nephrology (424)
- Neurology (3775)
- Nursing (207)
- Nutrition (560)
- Obstetrics and Gynecology (721)
- Occupational and Environmental Health (689)
- Oncology (1967)
- Ophthalmology (574)
- Orthopedics (233)
- Otolaryngology (301)
- Pain Medicine (248)
- Palliative Medicine (72)
- Pathology (469)
- Pediatrics (1092)
- Pharmacology and Therapeutics (455)
- Primary Care Research (442)
- Psychiatry and Clinical Psychology (3368)
- Public and Global Health (6447)
- Radiology and Imaging (1371)
- Rehabilitation Medicine and Physical Therapy (794)
- Respiratory Medicine (863)
- Rheumatology (394)
- Sexual and Reproductive Health (402)
- Sports Medicine (336)
- Surgery (434)
- Toxicology (51)
- Transplantation (185)
- Urology (164)
Thank you for visiting nature.com. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser (or turn off compatibility mode in Internet Explorer). In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript.
- View all journals
- Explore content
- About the journal
- Publish with us
- Sign up for alerts
- NATURE INDEX
- 17 November 2023
Hypotheses devised by AI could find ‘blind spots’ in research
- Matthew Hutson 0
Matthew Hutson is a science writer based in New York City.
You can also search for this author in PubMed Google Scholar

Credit: Olemedia/Getty
In early October, as the Nobel Foundation announced the recipients of this year’s Nobel prizes, a group of researchers, including a previous laureate, met in Stockholm to discuss how artificial intelligence (AI) might have an increasingly creative role in the scientific process. The workshop, led in part by Hiroaki Kitano, a biologist and chief executive of Sony AI in Tokyo, considered creating prizes for AIs and AI–human collaborations that produce world-class science. Two years earlier, Kitano proposed the Nobel Turing Challenge 1 : the creation of highly autonomous systems (‘AI scientists’) with the potential to make Nobel-worthy discoveries by 2050.
It’s easy to imagine that AI could perform some of the necessary steps in scientific discovery. Researchers already use it to search the literature, automate data collection, run statistical analyses and even draft parts of papers. Generating hypotheses — a task that typically requires a creative spark to ask interesting and important questions — poses a more complex challenge. For Sendhil Mullainathan, an economist at the University of Chicago Booth School of Business in Illinois, “it’s probably been the single most exhilarating kind of research I’ve ever done in my life”.
Network effects
AI systems capable of generating hypotheses go back more than four decades. In the 1980s, Don Swanson, an information scientist at the University of Chicago, pioneered literature-based discovery — a text-mining exercise that aimed to sift ‘undiscovered public knowledge’ from the scientific literature. If some research papers say that A causes B, and others that B causes C, for example, one might hypothesize that A causes C. Swanson created software called Arrowsmith that searched collections of published papers for such indirect connections and proposed, for instance, that fish oil, which reduces blood viscosity, might treat Raynaud’s syndrome, in which blood vessels narrow in response to cold 2 . Subsequent experiments proved the hypothesis correct.
Literature-based discovery and other computational techniques can organize existing findings into ‘knowledge graphs’, networks of nodes representing, say, molecules and properties. AI can analyse these networks and propose undiscovered links between molecule nodes and property nodes. This process powers much of modern drug discovery, as well as the task of assigning functions to genes. A review article published in Nature 3 earlier this year explores other ways in which AI has generated hypotheses, such as proposing simple formulae that can organize noisy data points and predicting how proteins will fold up. Researchers have automated hypothesis generation in particle physics, materials science, biology, chemistry and other fields.

An AI revolution is brewing in medicine. What will it look like?
One approach is to use AI to help scientists brainstorm. This is a task that large language models — AI systems trained on large amounts of text to produce new text — are well suited for, says Yolanda Gil, a computer scientist at the University of Southern California in Los Angeles who has worked on AI scientists. Language models can produce inaccurate information and present it as real, but this ‘hallucination’ isn’t necessarily bad, Mullainathan says. It signifies, he says, “‘here’s a kind of thing that looks true’. That’s exactly what a hypothesis is.”
Blind spots are where AI might prove most useful. James Evans, a sociologist at the University of Chicago, has pushed AI to make ‘alien’ hypotheses — those that a human would be unlikely to make. In a paper published earlier this year in Nature Human Behaviour 4 , he and his colleague Jamshid Sourati built knowledge graphs containing not just materials and properties, but also researchers. Evans and Sourati’s algorithm traversed these networks, looking for hidden shortcuts between materials and properties. The aim was to maximize the plausibility of AI-devised hypotheses being true while minimizing the chances that researchers would hit on them naturally. For instance, if scientists who are studying a particular drug are only distantly connected to those studying a disease that it might cure, then the drug’s potential would ordinarily take much longer to discover.
When Evans and Sourati fed data published up to 2001 to their AI, they found that about 30% of its predictions about drug repurposing and the electrical properties of materials had been uncovered by researchers, roughly six to ten years later. The system can be tuned to make predictions that are more likely to be correct but also less of a leap, on the basis of concurrent findings and collaborations, Evans says. But “if we’re predicting what people are going to do next year, that just feels like a scoop machine”, he adds. He’s more interested in how the technology can take science in entirely new directions.
Keep it simple
Scientific hypotheses lie on a spectrum, from the concrete and specific (‘this protein will fold up in this way’) to the abstract and general (‘gravity accelerates all objects that have mass’). Until now, AI has produced more of the former. There’s another spectrum of hypotheses, partially aligned with the first, which ranges from the uninterpretable (these thousand factors lead to this result) to the clear (a simple formula or sentence). Evans argues that if a machine makes useful predictions about individual cases — “if you get all of these particular chemicals together, boom, you get this very strange effect” — but can’t explain why those cases work, that’s a technological feat rather than science. Mullainathan makes a similar point. In some fields, the underlying principles, such as the mechanics of protein folding, are understood and scientists just want AI to solve the practical problem of running complex computations that determine how bits of proteins will move around. But in fields in which the fundamentals remain hidden, such as medicine and social science, scientists want AI to identify rules that can be applied to fresh situations, Mullainathan says.
In a paper presented in September 5 at the Economics of Artificial Intelligence Conference in Toronto, Canada, Mullainathan and Jens Ludwig, an economist at the University of Chicago, described a method for AI and humans to collaboratively generate broad, clear hypotheses. In a proof of concept, they sought hypotheses related to characteristics of defendants’ faces that might influence a judge’s decision to free or detain them before trial. Given mugshots of past defendants, as well the judges’ decisions, an algorithm found that numerous subtle facial features correlated with judges’ decisions. The AI generated new mugshots with those features cranked either up or down, and human participants were asked to describe the general differences between them. Defendants likely to be freed were found to be more “well-groomed” and “heavy-faced”. Mullainathan says the method could be applied to other complex data sets, such as electrocardiograms, to find markers of an impending heart attack that doctors might not otherwise know to look for. “I love that paper,” Evans says. “That’s an interesting class of hypothesis generation.”
In science, experimentation and hypothesis generation often form an iterative cycle: a researcher asks a question, collects data and adjusts the question or asks a fresh one. Ross King, a computer scientist at Chalmers University of Technology in Gothenburg, Sweden, aims to complete this loop by building robotic systems that can perform experiments using mechanized arms 6 . One system, called Adam, automated experiments on microbe growth. Another, called Eve, tackled drug discovery. In one experiment, Eve helped to reveal the mechanism by which a toothpaste ingredient called triclosan can be used to fight malaria.
Robot scientists
King is now developing Genesis, a robotic system that experiments with yeast. Genesis will formulate and test hypotheses related to the biology of yeast by growing actual yeast cells in 10,000 bioreactors at a time, adjusting factors such as environmental conditions or making genome edits, and measuring characteristics such as gene expression. Conceivably, the hypotheses could involve many subtle factors, but King says they tend to involve a single gene or protein whose effects mirror those in human cells, which would make the discoveries potentially applicable in drug development. King, who is on the organizing committee of the Nobel Turing Challenge, says that these “robot scientists” have the potential to be more consistent, unbiased, cheap, efficient and transparent than humans.
Researchers see several hurdles to and opportunities for progress. AI systems that generate hypotheses often rely on machine learning, which usually requires a lot of data. Making more papers and data sets openly available would help, but scientists also need to build AI that doesn’t just operate by matching patterns but can also reason about the physical world, says Rose Yu, a computer scientist at the University of California, San Diego. Gil agrees that AI systems should not be driven only by data — they should also be guided by known laws. “That’s a very powerful way to include scientific knowledge into AI systems,” she says.
As data gathering becomes more automated, Evans predicts that automating hypothesis generation will become increasingly important. Giant telescopes and robotic labs collect more measurements than humans can handle. “We naturally have to scale up intelligent, adaptive questions”, he says, “if we don’t want to waste that capacity.”
doi: https://doi.org/10.1038/d41586-023-03596-0
Kitano, H. npj Syst. Biol. Appl. 7 , 29 (2021).
Article PubMed Google Scholar
Swanson, D. R. Perspect. Biol. Med. 30 , 7–18 (1986).
Wang, H. et al. Nature 620 , 47–60 (2023).
Sourati, J. & Evans, J. A. Nature Hum. Behav. 7 , 1682–1696 (2023).
Ludwig, J. & Mullainathan, S. Working Paper 31017 (National Bureau of Economic Research, 2023).
King, R., Peter, O. & Courtney, P. in Artificial Intelligence in Science 129–139 (OECD Publishing, 2023).
Download references
Related Articles

- Machine learning
- Computer science
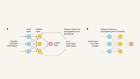
Physics solves a training problem for artificial neural networks
News & Views 07 AUG 24

These AI firms publish the world’s most highly cited work
News 01 AUG 24
Cheap light sources could make AI more energy efficient
News & Views 31 JUL 24

Fully forward mode training for optical neural networks
Article 07 AUG 24

Slow productivity worked for Marie Curie — here’s why you should adopt it, too
Career Feature 05 AUG 24

Quantum computing aims for diversity, one qubit at a time
Technology Feature 05 AUG 24

A publishing platform that places code front and centre
Technology Feature 07 AUG 24
China’s robotaxis need regulation
Correspondence 06 AUG 24
Faculty position – Department of Bone Marrow Transplantation & Cellular Therapy
Memphis, Tennessee
St. Jude Children's Research Hospital (St. Jude)
Faculty Position, Department of Diagnostic Imaging - AI Imaging Research
Neuroscience faculty position.
Neuroscience Faculty Position-Center for Pediatric Neurological Disease Research, St. Jude Pediatric Translational Neuroscience Initiative
RNA Mass Spectrometry Platform Manager (Level B)
The RNA mass spectrometry (MS) Platform Manager will be responsible for developing RNA MS workflows and managing the platform operation.
Monash University, Monash (AU)
Monash University
Postdoctoral Associate
Houston, Texas (US)
Baylor College of Medicine (BCM)
Sign up for the Nature Briefing newsletter — what matters in science, free to your inbox daily.
Quick links
- Explore articles by subject
- Guide to authors
- Editorial policies
- Open access
- Published: 29 August 2012
An automated framework for hypotheses generation using literature
- Vida Abedi 1 , 2 ,
- Ramin Zand 3 ,
- Mohammed Yeasin 1 , 2 &
- Fazle Elahi Faisal 1 , 2
BioData Mining volume 5 , Article number: 13 ( 2012 ) Cite this article
8526 Accesses
7 Citations
18 Altmetric
Metrics details
In bio-medicine, exploratory studies and hypothesis generation often begin with researching existing literature to identify a set of factors and their association with diseases, phenotypes, or biological processes. Many scientists are overwhelmed by the sheer volume of literature on a disease when they plan to generate a new hypothesis or study a biological phenomenon. The situation is even worse for junior investigators who often find it difficult to formulate new hypotheses or, more importantly, corroborate if their hypothesis is consistent with existing literature. It is a daunting task to be abreast with so much being published and also remember all combinations of direct and indirect associations. Fortunately there is a growing trend of using literature mining and knowledge discovery tools in biomedical research. However, there is still a large gap between the huge amount of effort and resources invested in disease research and the little effort in harvesting the published knowledge. The proposed hypothesis generation framework (HGF) finds “crisp semantic associations” among entities of interest - that is a step towards bridging such gaps.
Methodology
The proposed HGF shares similar end goals like the SWAN but are more holistic in nature and was designed and implemented using scalable and efficient computational models of disease-disease interaction. The integration of mapping ontologies with latent semantic analysis is critical in capturing domain specific direct and indirect “crisp” associations, and making assertions about entities (such as disease X is associated with a set of factors Z).
Pilot studies were performed using two diseases. A comparative analysis of the computed “associations” and “assertions” with curated expert knowledge was performed to validate the results. It was observed that the HGF is able to capture “crisp” direct and indirect associations, and provide knowledge discovery on demand.
Conclusions
The proposed framework is fast, efficient, and robust in generating new hypotheses to identify factors associated with a disease. A full integrated Web service application is being developed for wide dissemination of the HGF. A large-scale study by the domain experts and associated researchers is underway to validate the associations and assertions computed by the HGF.
Peer Review reports
The explosion of OMICS - based technologies, such as genomics, proteomics, and pharmaco-genomics, has generated a wave of information retrieval tools, such as SWAN [ 1 ], to mine the heterogeneous, high dimensional and large databases, as well as complex biological networks. The general characteristics of such complex systems as well as their robustness and dynamical properties were reported by many researchers (i.e., [ 2 , 3 ]). These reports of designing scalable and efficient knowledge discovery tools can further our understanding of complex biological systems. The burgeoning gap between the effort and investment made to acquire the knowledge about complexities of biological systems is disproportionately large compared to the development of knowledge discovery tools that can be used for effectively disseminating the acquired knowledge, generating and validating hypothesis, and understanding the complex causal relationships. Despite a plethora of efforts in reverse-engineering of complex systems to predict response to perturbations, there is a lack of significant effort to create a higher level abstraction of such complex biological systems using sources of information other than genetics data [ 2 , 4 ]. A high level view of complex systems would be very useful in generating new hypotheses and connecting seemingly unrelated entities. Such an abstraction could facilitate translational research and may prove vital in clinical studies by providing a valuable reference to the clinicians, researchers, and other domain experts.
Disease networks can provide a high level view of complex systems; however, the reported networks are mostly based on genetic and proteomic data [ 2 , 4 ]. Such networks could also be constructed based on literature data to incorporate a wider range of factors such as side effects and risk factors. Generating disease-models based on literature data is a very natural and efficient way to better understand and summarize the current knowledge about different high-level systems. A connection between two diseases can be formalized by risk factors, symptoms, treatment options, or any other diseases as compared to only common disease-genes. The relations between diseases can provide a systematic approach to identify missing links and potential associations. It will also create new avenues for collaborations and interdisciplinary research.
To construct a disease network based on literature data, it is imperative to have a scalable and efficient literature-mining tool to explore the huge textual resources. Nevertheless, mining of biological and medical literature is a very challenging task [ 5 – 7 ]. This can further be complicated by challenges with the implementation of relevant information extraction, also known as deep parsing, which is built on formal mathematical models. Deep parsing, also known as formal grammar, attempts to describe how text is generated in the human mind [ 5 ]. Deterministic or probabilistic context-free grammars are probably the most popular formal grammars [ 7 ]. Grammar-based information extraction techniques are computationally expensive as they require the evaluation of alternative ways to generate the same sentence. Grammar-based information could therefore be more precise but at the cost of reduced processing speed [ 5 ].
An alternative to the grammar-based methods are factorization methods such as Latent Semantic Analysis (LSA) [ 8 ], and Non-negative Matrix Factorization (NMF) [ 9 , 10 ]. Factorization methods rely on bag-of-word concept, and have therefore reduced computational complexity. LSA is a well known information retrieval technique which has been applied to many areas in bioinformatics. Arguably, LSA captures semantic relations between various concepts based on their distance in the reduced eigen space [ 11 ]. It has the advantage of extracting direct and indirect associations between entities. A commonly used distance measure in LSA is the cosine value of the angle between the document and query in the reduced eigen space.
Over the past two decades, medical text-mining has proved to be valuable in generating new exciting hypotheses. For instance, titles from MEDLINE were used to make connections between disconnected arguments: 1) the connection between migraine and magnesium deficiency [ 12 ] which has been verified experimentally; 2) between indomethacin and Alzheimer’s disease [ 12 ]; and finally 3) between Curcuma longa and retinal diseases [ 13 ]. Hypothesis generation in literature-mining relies on the fact that chance connections can emerge to be meaningful [ 7 ].
This paper designs and implements an efficient and scalable literature-mining framework to generate and also validate plausible hypotheses about various entities that include (but not limited to): risk factors, environmental factors, lifestyle, diseases, and disease groups. The proposed hypothesis generation framework (HGF) is implemented based on parameter optimized latent semantic analysis (POLSA) [ 14 ] and is suitable to capture direct and indirect association among concepts. It is easy to note that the overall performance and quality of results obtained through LSA-based systems is a function of the dictionary used. The concept of mapping ontologies was integrated with the POLSA to overcome such limitations and to provide crisp associations. In particular, the Medical Subject Headings (MESH) is used to construct the dictionary. Such a dictionary allows a more efficient use of the LSA technique in finding semantically related entities in the biological and medical sciences. This framework can be used to generate customized disease-disease interaction networks, to facilitate interdisciplinary collaborations between scientists and organizations, to discover hidden knowledge, and to spawn new research directions. In addition, the concept of statistical disease modeling was introduced to compute the strongly related, related, and not related concepts.
The following section describes the proposed hypothesis generation framework and its evaluation. Two case studies were performed to showcase the potential and utility of the proposed method. Finally, the paper ends with a brief conclusion and discussions about the strengths and weaknesses of the method.
Results and discussion
Hypotheses generation framework (hgf).
The HGF has three major modules: Ontology Mapping to generate data-driven domain specific dictionaries, a parameter optimized latent semantic analysis (POLSA), and Disease Model. The schematic diagram of the overall HGF framework is shown in the Figure 1 (A). The model is constructed using the POLSA framework, and it is based on the selected documents and the dictionary (Figure 1 C). Users can query the model and the output is a ranked list of headings. These ranked headings are grouped into three sets (unknown factors, potential factors, or established factors) using the Disease Model module (Figure 1 C and 1 D). Analyzing the headings in the three sets can facilitate hypothesis generation and information retrieval based on user query.
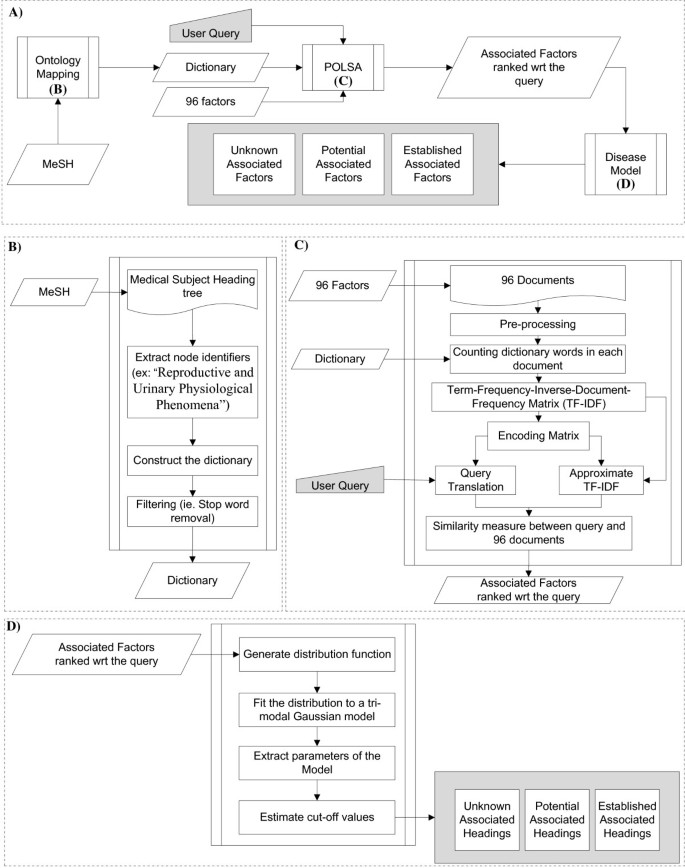
Flow diagram of the hypothesis generation framework (HGF). A ) In a medical and biological setting, Ontology Mapping could use the Medical Subject Heading (MeSH) and generate a context specific dictionary, which is one of the parameters of the POLSA model. Associated factors are ranked based on a User Query which can be any word(s) in the dictionary. These factors are subsequently grouped into three different bins (unknown factors, potential factors or established factors) based on our Disease Model. B ) Ontology Mapping to create domain specific dictionary. C ) Parameter Optimized Latent Semantic Analysis Module. D ) Disease Model Module.
Ontology mapping
MeSH is used to generate the dictionary in the POLSA model. The mapping of MeSH ontology to create the dictionary for the POLSA significantly enhances the quality of results and provides a crisp association of semantically related entities in biological and medical science. All MeSH headings are reduced to single words to create the context specific and data driven dictionary (see Figure 1 B). For instance, “Reproductive and Urinary Physiological Phenomena” is a MeSH term and is reduced to five words in the dictionary (1. Reproductive, 2. and, 3. Urinary, 4. Physiological, and 5. Phenomena). In the filtering step, duplicates as well as stop words such as “and” or words containing fewer than three characters are removed. The final size of this dictionary is 19,165 words. Any dictionary word could be used as a query to the HGF. For instance, the disease “stroke” is a query in this study. The highly ranked factors with respect to a query-disease are considered factors associated with that disease. Cosine similarity measure is used as a metric in the HGF.
POLSA module
In order to develop an effective literature-mining framework to model disease-disease interaction networks, generate plausible new hypotheses, and support knowledge-discovery by finding semantically related entities, a Parameter Optimized LSA (POLSA) [ 14 ] was re-designed and adopted in the proposed HGF framework.
In addition, a set of associated factors was selected to represent interaction between diseases. Ninety-six common associated factors (see Table 1 ) were selected through a literature review from numerous medical articles by two domain experts. As the first step, a set of articles was selected by querying the PubMed database using a series of diseases and factors. In the second step, the retrieved articles were manually reviewed by domain experts and entities that were associated with diseases or factors were selected. All articles considered for this analysis were peer reviewed articles. In addition, some common diseases such as diabetes and depression were also included in the set of 96 factors, as these are believed to be, in many instances, risk factors to other diseases. Therefore, the set of 96 associated factors represents a wide range of factors including generic factors such as depression and infection as well as specific factors such as vitamin E. As the final step, the set was further revised by an expert in the medical field. Using the improved POLSA technique [ 14 ], meaningful associations from the textual data in the PubMed database are extracted and mined. Furthermore, the factors are ranked based on their level of association to a given query.
Titles and abstracts from PubMed (for the past twenty years) for each of the 96 factors were downloaded in a local machine. On average there were 47,570 abstracts per factor; the specific factors such as “maternal influenza” had fewer abstracts associated with them (minimum of 160 abstracts/factor) and the more generic factors such as “hormone” were associated with a greater number of abstracts (a maximum of 557,554 abstracts/factor). The complete collection was then used to construct the knowledge space for the POLSA model. Using a query such as “Parkinson” or “stroke” the 96 factors were then ranked based on their relative level of associations to the query. The distribution of a set of associated factors with respect to a disease was modeled as a tri-modal distribution: a distribution which has three modes. This is due to the fact that some factors are known to be associated with the disease and have high scores. Similarly, some factors are known to be unassociated to the disease and these have negative scores; in addition, some factors may or may not be associated to the disease and these have low similarity scores. Matlab was used to generate two tri-modal distributions based on general Gaussian models for the two distributions obtained from queries “stroke” and “Parkinson”. The model uses the following formulation to describe the tri-modal Gaussian distribution:
Where α 1 , α 2 and α 3 are the scaling factors; μ 1 , μ 2 and μ 3 are the position of the center of the peaks, and σ 1 , σ 2 , σ 3 control the width of the distributions. The goodness of fit was measured using an R-square score.
- Disease model
Using a disease model (see Figure 2 ), it was possible to map the mixture of three Gaussian distributions into easy to understandable categories. The implicit assumption is that if associated factors of a disease are well known, a large body of literature will be available to corroborate the existence of such associations. On the other hand, if associated factors of a disease are not well documented, the factors are weakly associated to the disease with few factors displaying a high level of association (Disease X versus Disease Y as shown in the Figure 2 ). Since the distribution of association level of factors (including risk factors) will be different in the two scenarios. In the first case (Disease Y) the two dominating distributions are the factors that are associated and those that are not associated with the disease; in the second case (Disease X) the dominating distribution is that of the potential factors. In essence, if one accepts this assumption then the distribution of associated factors follows a tri-modal distribution and it will be intuitive to measure the level of association for different factors with respect to a given disease. Utilization of a disease model (by a tri-modal distribution) allows better identification of the three sets of factors: unknown associations, potential associations and established associations.
Model for the distribution of associated factors of a given disease. If associated factors – such as risk factors – of a disease are well known as in the case for Disease Y, then the two dominating distributions are the factors that are associated and those that are not associated with the disease; if on other hand the associated factors of a disease are not well documented (Disease X) then the dominating distribution is that of the potential factors.
Separating the three distributions allows implementation of a dynamic and data-driven threshold calculation. Hence, the parameters of the distributions can be used to model a cut-off threshold for the factors that are established, potential, or unknown. This method is empirical and provides an intuitive approach to evaluate the results. The score can be further optimized in a heuristic manner with utilization of a large-scale and comprehensive ground truth set. Furthermore, the highly associated factors to the disease are the well known factors; the hidden knowledge on the other hand resides in the region where the associations are positive yet weak.
Model evaluation
Two diseases, namely, Ischemic Stroke (IS) and Parkinson’s Disease (PD), were used as queries to the hypothesis generation system. The distribution of associated factors is presented in the Figure 3 . The results were compared with MedLink neurology [ 15 ], a web resource used by clinicians. Comparative results were summarized in the Figure 4 . In the case of IS, most of the associated factors are identified by both systems; however there is a set of factors that have only been identified by the proposed approach. In the case of the PD, a large number of factors have been identified by both systems. However, there are a number of factors that have only been identified by the proposed HGF and only a handful that are mentioned in the MedLink neurology which have positive but low similarity score in the hypothesis generation framework.
Number of factors identified by MedLink Neurology and by HGF for IS and PD. Association levels for IS measured by HGF are high (0.3 < cosine score) and possible (0.1 < cosine score < 0.3); association levels for PD measured by HGF are high (0.2 < cosine score), possible (0.1 < cosine score < 0.2) or low (0.05 < cosine score < 0.1).
Distribution of similarity score (dashed line) for risk factors associated with IS and PD. The frequency represents the number of factors at each cosine similarity level (−1 to +1). Tri-modal distribution models are represented by solid lines.
The tri-modal distribution model is used to group the associated factors into different levels. The cut-off values to differentiate between different association levels vary slightly depending on the distribution of the similarity scores. The ideal decision boundary can be found if a large number of ground truth cases are available; in this situation the decision boundary is selected intuitively based on the shape of the distributions. For example, in the case of IS, factors are considered highly associated if their cosine score is greater than 0.3, factors are possible associated if their score is between 0.1 and 0.3 and are possibly not associated if their score is lower than 0.1. In the case of PD, factors are considered highly associated if their cosine score is greater than 0.2, factors are possibly associated if their cosine score is between 0.1 and 0.2 and finally the factors with scores between 0.05 and 0.1 are considered associated at low level, factors with scores lower than 0.05 are considered possibly not associated with the Parkinson’s Disease.
In the case of IS, the distribution of known associated factors are more shifted to the right as compared to the factors in PD, hence the separation between the known and unknown factors is more pronounced. In addition to that, associations at both extreme levels (close to +1.0 and −1.0) are likely to be common knowledge; however, the hidden knowledge tends to be captured at similarity scores that are low yet positive. Nonetheless, it is not realistic to compare the precise similarity score values in order to give more importance to one factor versus another factor mainly because there is a systemic bias that is inherent to the biological text data and causes the generic factors to be an underestimate of the true value (data not shown); hence a direct comparison would fail in this case if no additional normalization steps are taken.
Figure 3 summarizes a comparative analysis of MedLink Neurology and HGF for IS and PD. Overall in the case of IS, twelve factors were identified by both systems and six factors were identified by the HGF. In the case of PD, twelve factors were identified by both systems, ten factors were identified by the HGF and five factors were identified by MedLink Neurology. But, these factors had a low association level in HGF. The five factors were either very generic or were not exactly mapped in the set of the 96 factors, hence a direct comparison could not be made. Finally, this small scale comparative analysis corroborates the hypothesis that HGF based on literature can better predict the associated factors for diseases such as IS when the risk and associated factors are well studied and documented. In both cases, MedLink, Neurology, and HGF predicted twelve common associated factors; however, in the case of PD ten new factors were predicted in comparison to six in the case of IS.
De novo hypothesis generation can provide an approach on how we design experiments and select the parameters for the study. Interestingly, associations detected by the proposed framework can facilitate extraction of interesting observations and new trends in the field. For instance, it was found that PD could possibly be associated with immunological disorders; this is an intriguing observation. This analysis also facilitates interdisciplinary research and enhances interaction among scientists from sub-specialized fields. A manual review of the literature is performed to find evidences for some of the associations found only by the HGF; Table 2 summarizes these results.
There are three main limitations in the presented framework. We are currently in the process of finding solutions for these limitations. 1) Manual selection of the factors creates bias in the dataset and also limits its scalability property. To alleviate this problem, MeSH hierarchy will be used to generate the set of factors. MeSH comprises more than 25,000 subjects headings organized in an eleven-level hierarchy. 2) In the set of 96 factors, some factors were very generic and some very specific, therefore, there was a systemic bias in the dataset which caused the score for generic factors to be an underestimate of the true values and factors with limited information to be overestimated (data not shown). To partially solve this technical difficulty, an improved method based on local LSA is being developed in our lab. And finally, 3) looking only at literature from the past twenty years was not sufficient for the HGF. The expansion of the literature is necessary based on the observation that the association between head trauma and PD was significantly lower than expected.
Generating new hypotheses by mining a vast amount of raw unstructured knowledge from the archived reported literature may help in identifying new research trends as well as promoting interdisciplinary studies. In addition, the presented framework is not limited to uncovering disease-disease interactions; any word from MeSH can be used to query the system, and its associated factors can be identified accordingly. Disease-disease interaction networks, interaction networks among chemical compounds, drug-drug interaction networks, or any specific type of interaction network can be constructed using the HGF. The common basis for all these networks is the knowledge embedded in the literature. Application of this framework is broad as its usage is not limited to any specific domain. For instance, uncovering drug-drug interactions is valuable in drug development and drug administration, uncovering disease-disease interaction is important in understanding disease mechanism’s and advancing biology through integrated interdisciplinary research. Even though the framework is not limited to diseases, in this study two neurological diseases were used to test the system and demonstrate the power and applicability of the framework.
In addition to addressing the limitations of the framework, work is in progress to expand the HGF framework to allow the user to generate disease networks based on a number of user-defined queries. Such customized networks can be valuable to a wide range of scientists by promoting a faster identification of associated factors and detection of disease-disease interactions. Disease networks based on genetics and proteomics data display many connections between individual disorders and disease categories [ 2 , 4 ]. Therefore, as expected each human disorder does not seem to have unique origins or be independent of other disorders. To uncover potential links between two disorders knowledge extraction from medical literature could be greatly beneficial and reliable.

Authors’ information
VA is a Ph.D. candidate in Electrical and Computer Engineering at the University of Memphis; she has a B.A.Sc. in Computer Engineering and B.Sc. in Biochemistry in addition to a M.Sc. in Cellular Molecular Medicine and a second M.Sc. in Bioinformatics. Her research interests are interdisciplinary research in Medical Informatics and Systems Biology. VA’s research incorporates a systems approach to understanding gene regulatory networks, which combines mathematical modeling and molecular biology wet lab techniques. Her recent contributions are in medical informatics where her board understanding of interdisciplinary issues as well as deep knowledge in mathematics and experimental biology are fundamental in designing and performing experiments in translational research.
RZ is a M.D. in the department of Neurology at the University of Tennessee. He also holds a Masters of Public Health. His research interests include Vascular Neurology and Bioinformatics. Over the past few years, RZ has contributed to bridge the gap between clinical findings and application of bioinformatics tools.
FEF is a PhD candidate in Electrical and Computer Engineering at The University of Memphis; he has a B.Sc. in Computer Science and Engineering, M.Sc. degree in Computer Science and Engineering and a second M.Sc. degree in Bioinformatics. His research interests are biological information retrieval and data mining. FEF possesses good knowledge in software design and development. He participated in software development of some national and international research projects, such as Codewitz Asia-Link Project of European Union.
MY is an Associate Professor in the department of Electrical and Computer Engineering, adjunct faculty member of Biomedical Engineering and Bioinformatics Program, and an affiliated member of the Institute for Intelligent Systems (IIS) at The University of Memphis (U of M). He is a senior member of the IEEE. He made significant contributions in the research and development of real-time computer vision solutions for academic research and commercial applications. He has been involved with several technological innovations, including classifying gender, age group, ethnicity and emotion, face detection, recognition of human activities in video, and speech-gesture enabled sophisticated natural human-computer interfaces. Some of his research on facial image analysis and hand gesture recognition is used in developing several commercial products by the Videomining Inc.
Abbreviations
Hypothesis generation framework
Ischemic stroke
Latent semantic analysis
Medical subject heading
Non-negative matrix factorization
Parkinson’s disease
Parameter optimized latent semantic analysis.
Gao Y, Kinoshita J, Wu E, Miller E, Lee R, Seaborne A, Cayzer S, Clark T: SWAN: A Distributed Knowledge Infrastructure for Alzheimer Disease Research. Journal of Web Semantics. 2006, 4 (3): 222-228. 10.1016/j.websem.2006.05.006.
Article Google Scholar
Goh KI, Cusick ME, Valle D, Childs B, Vidal M, Barabási AL: The human disease network. Proc Natl Acad Sci USA. 2007, 104 (21): 8685-8690. 10.1073/pnas.0701361104.
Article CAS PubMed PubMed Central Google Scholar
Ideker T, Thorsson V, Ranish JA, Christmas R, Buhler J, Eng JK, Bumgarner R, Goodlett DR, Aebersold R, Hood L: Integrated genomic and proteomic analyses of a systematically perturbed metabolic network. Science. 2001, 292 (5518): 929-934. 10.1126/science.292.5518.929.
Article CAS PubMed Google Scholar
Zhang X, Zhang R, Jiang Y, Sun P, Tang G, Wang X, Lv H, Li X: The expanded human disease network combining protein–protein interaction information. Eur J Hum Genet. 2011, 19 (7): 783-788. 10.1038/ejhg.2011.30.
Rzhetsky A, Seringhaus M, Gerstein M: Seeking a new biology through text mining. Cell. 2008, 134 (1): 9-13. 10.1016/j.cell.2008.06.029.
Hirschman L, Morgan AA, Yeh AS: Rutabaga by any other name: extracting biological names. J Biomed Inform. 2002, 35 (4): 247-259. 10.1016/S1532-0464(03)00014-5.
Wilbur WJ, Hazard GF, Divita G, Mork JG, Aronson AR, Browne AC: Analysis of biomedical text for chemical names: a comparison of three methods. Proc AMIA Symp. 1999, 176-180. http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2232672/ .
Landauer TK, Dumais ST: A solution to plato’s problem: the latent semantic analysis theory of the acquisition, induction, and representation of knowledge. Psychol Rev. 1997, 104: 211-240.
Lee DD, Seung HS: Learning the parts of objects by non-negative matrix factorization. Nature. 1999, 401: 788-791. 10.1038/44565.
Paatero P, Tapper U: Positive matrix factorization: a non-negative factor model with optimal utilization of error estimates of data values. Environmetrics. 1994, 5: 111-126. 10.1002/env.3170050203.
Berry MW, Browne M: Understanding Search Engines: Mathematical Modeling and Text Retrieval. 1990, Philadelphia, USA: SIAM
Google Scholar
Swanson D, Smalheiser N: Assessing a gap in the biomedical literature: magnesium deficiency and neurologic disease. Neurosci Res Commun. 1994, 15: 1-9.
Srinivasan P, Libbus B: Mining MEDLINE for implicit links between dietary substances and diseases. Bioinformatics. 2004, 20 (Suppl 1): i290-i296. 10.1093/bioinformatics/bth914.
Yeasin M, Malempati H, Homayouni R, Sorower MS: A systematic study on latent semantic analysis model parameters for mining biomedical literature. Conference Proceedings: BMC Bioinformatics. 2009, 10 (Suppl. 7): A6-
Medlink Neurology. [ http://www.medlink.com/medlinkcontent.asp ]
Catling LA, Abubakar I, Lake IR, Swift L, Hunter PR: A systematic review of analytical observational studies investigating the association between cardiovascular disease and drinking water hardness. J Water Health. 2008, 6 (4): 433-442. 10.2166/wh.2008.054.
Article PubMed Google Scholar
Menown IA, Shand JA: Recent advances in cardiology. Future Cardiol. 2010, 6 (1): 11-17. 10.2217/fca.09.59.
Tafet GE, Idoyaga-Vargas VP, Abulafia DP, Calandria JM, Roffman SS, Chiovetta A, Shinitzky M: Correlation between cortisol level and serotonin uptake in patients with chronic stress and depression. Cogn Affect Behav Neurosci. 2001, 1 (4): 388-393. 10.3758/CABN.1.4.388.
Williams GP: The role of oestrogen in the pathogenesis of obesity, type 2 diabetes, breast cancer and prostate disease. Eur J Cancer Prev. 2010, 19 (4): 256-271. 10.1097/CEJ.0b013e328338f7d2.
Schürks M, Glynn RJ, Rist PM, Tzourio C, Kurth T: Effects of vitamin E on stroke subtypes: meta-analysis of randomised controlled trials. BMJ. 2010, 341: c5702-10.1136/bmj.c5702.
Article PubMed PubMed Central Google Scholar
Benkler M, Agmon-Levin N, Shoenfeld Y: Parkinson’s disease, autoimmunity, and olfaction. Int J Neurosci. 2009, 119 (12): 2133-2143. 10.3109/00207450903178786.
Moscavitch SD, Szyper-Kravitz M, Shoenfeld Y: Autoimmune pathology accounts for common manifestations in a wide range of neuro-psychiatric disorders: the olfactory and immune system interrelationship. Clin Immunol. 2009, 130 (3): 235-243. 10.1016/j.clim.2008.10.010.
Faria AM, Weiner HL: Oral tolerance. Immunol Rev. 2005, 206: 232-259. 10.1111/j.0105-2896.2005.00280.x.
Teixeira G, Paschoal PO, de Oliveira VL, Pedruzzi MM, Campos SM, Andrade L, Nobrega A: Diet selection in immunologically manipulated mice. Immunobiology. 2008, 213 (1): 1-12. 10.1016/j.imbio.2007.08.001.
Schiffman SS, Sattely-Miller EA, Taylor EL, Graham BG, Landerman LR, Zervakis J, Campagna LK, Cohen HJ, Blackwell S, Garst JL: Combination of flavor enhancement and chemosensory education improves nutritional status in older cancer patients. J Nutr Health Aging. 2007, 11 (5): 439-454.
CAS PubMed Google Scholar
Murphy C, Davidson TM, Jellison W, Austin S, Mathews WC, Ellison DW, Schlotfeldt C: Sinonasal disease and olfactory impairment in HIV disease: endoscopic sinus surgery and outcome measures. Laryngoscope. 2000, 110 (10 Pt 1): 1707-1710.
Zucco GM, Ingegneri G: Olfactory deficits in HIV-infected patients with and without AIDS dementia complex. Physiol Behav. 2004, 80 (5): 669-674. 10.1016/j.physbeh.2003.12.001.
Tandeter H, Levy A, Gutman G, Shvartzman P: Subclinical thyroid disease in patients with Parkinson’s disease. Arch Gerontol Geriatr. 2001, 33 (3): 295-300. 10.1016/S0167-4943(01)00196-0.
Chinnakkaruppan A, Das S, Sarkar PK: Age related and hypothyroidism related changes on the stoichiometry of neurofilament subunits in the developing rat brain. Int J Dev Neurosci. 2009, 27 (3): 257-261. 10.1016/j.ijdevneu.2008.12.007.
García-Moreno JM, Chacón-Peña J: Hypothyroidism and Parkinson’s disease and the issue of diagnostic confusion. Mov Disord. 2003, 18 (9): 1058-1059. 10.1002/mds.10475.
Munhoz RP, Teive HA, Troiano AR, Hauck PR, Herdoiza Leiva MH, Graff H, Werneck LC: Parkinson’s disease and thyroid dysfunction. Parkinsonism Relat Disord. 2004, 10 (6): 381-383. 10.1016/j.parkreldis.2004.03.008.
Ferreira JJ, Neutel D, Mestre T, Coelho M, Rosa MM, Rascol O, Sampaio C: Skin cancer and Parkinson’s disease. Mov Disord. 2010, 25 (2): 139-148. 10.1002/mds.22855.
Download references
Acknowledgements
This work was supported by the Electrical and Computer Engineering Department and Bioinformatics Program at the University of Memphis, by the University of Tennessee Health Science Center (UTHSC), as well as by NSF grant NSF-IIS-0746790. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the funding institution.
Author information
Authors and affiliations.
Department of Electrical and Computer Engineering, Memphis University, Memphis, TN, 38152, USA
Vida Abedi, Mohammed Yeasin & Fazle Elahi Faisal
College of Arts and Sciences, Bioinformatics Program, Memphis University, Memphis, TN, 38152, USA
Department of Neurology, University of Tennessee Health Science Center, Memphis, TN, 38163, USA
You can also search for this author in PubMed Google Scholar
Corresponding author
Correspondence to Mohammed Yeasin .
Additional information
Competing interests.
The authors declare that they have no competing interests.
Authors’ contributions
VA designed and carried out the experiments, participated in the development of the methods, analyzed the results and drafted the manuscript. RZ participated in the development of the methods, designed the validation experiments for the two test cases and reviewed the manuscript. FEF participated in the implementation of the algorithms. MY participated in the development of the methods, supervised the experiments and edited the manuscript. All authors have read, and approved the final version of the manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Authors’ original file for figure 1
Authors’ original file for figure 2, authors’ original file for figure 3, authors’ original file for figure 4, rights and permissions.
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Reprints and permissions
About this article
Cite this article.
Abedi, V., Zand, R., Yeasin, M. et al. An automated framework for hypotheses generation using literature. BioData Mining 5 , 13 (2012). https://doi.org/10.1186/1756-0381-5-13
Download citation
Received : 30 March 2012
Accepted : 13 July 2012
Published : 29 August 2012
DOI : https://doi.org/10.1186/1756-0381-5-13
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Disease network
- Biological literature-mining
- Hypothesis generation
- Knowledge discovery
- MeSH ontology
BioData Mining
ISSN: 1756-0381
- General enquiries: [email protected]
- Search Menu
Sign in through your institution
- Browse content in A - General Economics and Teaching
- Browse content in A1 - General Economics
- A11 - Role of Economics; Role of Economists; Market for Economists
- Browse content in B - History of Economic Thought, Methodology, and Heterodox Approaches
- Browse content in B4 - Economic Methodology
- B49 - Other
- Browse content in C - Mathematical and Quantitative Methods
- Browse content in C0 - General
- C00 - General
- C01 - Econometrics
- Browse content in C1 - Econometric and Statistical Methods and Methodology: General
- C10 - General
- C11 - Bayesian Analysis: General
- C12 - Hypothesis Testing: General
- C13 - Estimation: General
- C14 - Semiparametric and Nonparametric Methods: General
- C18 - Methodological Issues: General
- Browse content in C2 - Single Equation Models; Single Variables
- C21 - Cross-Sectional Models; Spatial Models; Treatment Effect Models; Quantile Regressions
- C23 - Panel Data Models; Spatio-temporal Models
- C26 - Instrumental Variables (IV) Estimation
- Browse content in C3 - Multiple or Simultaneous Equation Models; Multiple Variables
- C30 - General
- C31 - Cross-Sectional Models; Spatial Models; Treatment Effect Models; Quantile Regressions; Social Interaction Models
- C32 - Time-Series Models; Dynamic Quantile Regressions; Dynamic Treatment Effect Models; Diffusion Processes; State Space Models
- C35 - Discrete Regression and Qualitative Choice Models; Discrete Regressors; Proportions
- Browse content in C4 - Econometric and Statistical Methods: Special Topics
- C40 - General
- Browse content in C5 - Econometric Modeling
- C52 - Model Evaluation, Validation, and Selection
- C53 - Forecasting and Prediction Methods; Simulation Methods
- C55 - Large Data Sets: Modeling and Analysis
- Browse content in C6 - Mathematical Methods; Programming Models; Mathematical and Simulation Modeling
- C63 - Computational Techniques; Simulation Modeling
- C67 - Input-Output Models
- Browse content in C7 - Game Theory and Bargaining Theory
- C71 - Cooperative Games
- C72 - Noncooperative Games
- C73 - Stochastic and Dynamic Games; Evolutionary Games; Repeated Games
- C78 - Bargaining Theory; Matching Theory
- C79 - Other
- Browse content in C8 - Data Collection and Data Estimation Methodology; Computer Programs
- C83 - Survey Methods; Sampling Methods
- Browse content in C9 - Design of Experiments
- C90 - General
- C91 - Laboratory, Individual Behavior
- C92 - Laboratory, Group Behavior
- C93 - Field Experiments
- C99 - Other
- Browse content in D - Microeconomics
- Browse content in D0 - General
- D00 - General
- D01 - Microeconomic Behavior: Underlying Principles
- D02 - Institutions: Design, Formation, Operations, and Impact
- D03 - Behavioral Microeconomics: Underlying Principles
- D04 - Microeconomic Policy: Formulation; Implementation, and Evaluation
- Browse content in D1 - Household Behavior and Family Economics
- D10 - General
- D11 - Consumer Economics: Theory
- D12 - Consumer Economics: Empirical Analysis
- D13 - Household Production and Intrahousehold Allocation
- D14 - Household Saving; Personal Finance
- D15 - Intertemporal Household Choice: Life Cycle Models and Saving
- D18 - Consumer Protection
- Browse content in D2 - Production and Organizations
- D20 - General
- D21 - Firm Behavior: Theory
- D22 - Firm Behavior: Empirical Analysis
- D23 - Organizational Behavior; Transaction Costs; Property Rights
- D24 - Production; Cost; Capital; Capital, Total Factor, and Multifactor Productivity; Capacity
- Browse content in D3 - Distribution
- D30 - General
- D31 - Personal Income, Wealth, and Their Distributions
- D33 - Factor Income Distribution
- Browse content in D4 - Market Structure, Pricing, and Design
- D40 - General
- D41 - Perfect Competition
- D42 - Monopoly
- D43 - Oligopoly and Other Forms of Market Imperfection
- D44 - Auctions
- D47 - Market Design
- D49 - Other
- Browse content in D5 - General Equilibrium and Disequilibrium
- D50 - General
- D51 - Exchange and Production Economies
- D52 - Incomplete Markets
- D53 - Financial Markets
- D57 - Input-Output Tables and Analysis
- Browse content in D6 - Welfare Economics
- D60 - General
- D61 - Allocative Efficiency; Cost-Benefit Analysis
- D62 - Externalities
- D63 - Equity, Justice, Inequality, and Other Normative Criteria and Measurement
- D64 - Altruism; Philanthropy
- D69 - Other
- Browse content in D7 - Analysis of Collective Decision-Making
- D70 - General
- D71 - Social Choice; Clubs; Committees; Associations
- D72 - Political Processes: Rent-seeking, Lobbying, Elections, Legislatures, and Voting Behavior
- D73 - Bureaucracy; Administrative Processes in Public Organizations; Corruption
- D74 - Conflict; Conflict Resolution; Alliances; Revolutions
- D78 - Positive Analysis of Policy Formulation and Implementation
- Browse content in D8 - Information, Knowledge, and Uncertainty
- D80 - General
- D81 - Criteria for Decision-Making under Risk and Uncertainty
- D82 - Asymmetric and Private Information; Mechanism Design
- D83 - Search; Learning; Information and Knowledge; Communication; Belief; Unawareness
- D84 - Expectations; Speculations
- D85 - Network Formation and Analysis: Theory
- D86 - Economics of Contract: Theory
- D89 - Other
- Browse content in D9 - Micro-Based Behavioral Economics
- D90 - General
- D91 - Role and Effects of Psychological, Emotional, Social, and Cognitive Factors on Decision Making
- D92 - Intertemporal Firm Choice, Investment, Capacity, and Financing
- Browse content in E - Macroeconomics and Monetary Economics
- Browse content in E0 - General
- E00 - General
- E01 - Measurement and Data on National Income and Product Accounts and Wealth; Environmental Accounts
- E02 - Institutions and the Macroeconomy
- E03 - Behavioral Macroeconomics
- Browse content in E1 - General Aggregative Models
- E10 - General
- E12 - Keynes; Keynesian; Post-Keynesian
- E13 - Neoclassical
- Browse content in E2 - Consumption, Saving, Production, Investment, Labor Markets, and Informal Economy
- E20 - General
- E21 - Consumption; Saving; Wealth
- E22 - Investment; Capital; Intangible Capital; Capacity
- E23 - Production
- E24 - Employment; Unemployment; Wages; Intergenerational Income Distribution; Aggregate Human Capital; Aggregate Labor Productivity
- E25 - Aggregate Factor Income Distribution
- Browse content in E3 - Prices, Business Fluctuations, and Cycles
- E30 - General
- E31 - Price Level; Inflation; Deflation
- E32 - Business Fluctuations; Cycles
- E37 - Forecasting and Simulation: Models and Applications
- Browse content in E4 - Money and Interest Rates
- E40 - General
- E41 - Demand for Money
- E42 - Monetary Systems; Standards; Regimes; Government and the Monetary System; Payment Systems
- E43 - Interest Rates: Determination, Term Structure, and Effects
- E44 - Financial Markets and the Macroeconomy
- Browse content in E5 - Monetary Policy, Central Banking, and the Supply of Money and Credit
- E50 - General
- E51 - Money Supply; Credit; Money Multipliers
- E52 - Monetary Policy
- E58 - Central Banks and Their Policies
- Browse content in E6 - Macroeconomic Policy, Macroeconomic Aspects of Public Finance, and General Outlook
- E60 - General
- E62 - Fiscal Policy
- E66 - General Outlook and Conditions
- Browse content in E7 - Macro-Based Behavioral Economics
- E71 - Role and Effects of Psychological, Emotional, Social, and Cognitive Factors on the Macro Economy
- Browse content in F - International Economics
- Browse content in F0 - General
- F00 - General
- Browse content in F1 - Trade
- F10 - General
- F11 - Neoclassical Models of Trade
- F12 - Models of Trade with Imperfect Competition and Scale Economies; Fragmentation
- F13 - Trade Policy; International Trade Organizations
- F14 - Empirical Studies of Trade
- F15 - Economic Integration
- F16 - Trade and Labor Market Interactions
- F18 - Trade and Environment
- Browse content in F2 - International Factor Movements and International Business
- F20 - General
- F21 - International Investment; Long-Term Capital Movements
- F22 - International Migration
- F23 - Multinational Firms; International Business
- Browse content in F3 - International Finance
- F30 - General
- F31 - Foreign Exchange
- F32 - Current Account Adjustment; Short-Term Capital Movements
- F34 - International Lending and Debt Problems
- F35 - Foreign Aid
- F36 - Financial Aspects of Economic Integration
- Browse content in F4 - Macroeconomic Aspects of International Trade and Finance
- F40 - General
- F41 - Open Economy Macroeconomics
- F42 - International Policy Coordination and Transmission
- F43 - Economic Growth of Open Economies
- F44 - International Business Cycles
- Browse content in F5 - International Relations, National Security, and International Political Economy
- F50 - General
- F51 - International Conflicts; Negotiations; Sanctions
- F52 - National Security; Economic Nationalism
- F55 - International Institutional Arrangements
- Browse content in F6 - Economic Impacts of Globalization
- F60 - General
- F61 - Microeconomic Impacts
- F62 - Macroeconomic Impacts
- F63 - Economic Development
- Browse content in G - Financial Economics
- Browse content in G0 - General
- G00 - General
- G01 - Financial Crises
- G02 - Behavioral Finance: Underlying Principles
- Browse content in G1 - General Financial Markets
- G10 - General
- G11 - Portfolio Choice; Investment Decisions
- G12 - Asset Pricing; Trading volume; Bond Interest Rates
- G14 - Information and Market Efficiency; Event Studies; Insider Trading
- G15 - International Financial Markets
- G18 - Government Policy and Regulation
- G19 - Other
- Browse content in G2 - Financial Institutions and Services
- G20 - General
- G21 - Banks; Depository Institutions; Micro Finance Institutions; Mortgages
- G22 - Insurance; Insurance Companies; Actuarial Studies
- G23 - Non-bank Financial Institutions; Financial Instruments; Institutional Investors
- G24 - Investment Banking; Venture Capital; Brokerage; Ratings and Ratings Agencies
- G28 - Government Policy and Regulation
- Browse content in G3 - Corporate Finance and Governance
- G30 - General
- G31 - Capital Budgeting; Fixed Investment and Inventory Studies; Capacity
- G32 - Financing Policy; Financial Risk and Risk Management; Capital and Ownership Structure; Value of Firms; Goodwill
- G33 - Bankruptcy; Liquidation
- G34 - Mergers; Acquisitions; Restructuring; Corporate Governance
- G38 - Government Policy and Regulation
- Browse content in G4 - Behavioral Finance
- G40 - General
- G41 - Role and Effects of Psychological, Emotional, Social, and Cognitive Factors on Decision Making in Financial Markets
- Browse content in G5 - Household Finance
- G50 - General
- G51 - Household Saving, Borrowing, Debt, and Wealth
- Browse content in H - Public Economics
- Browse content in H0 - General
- H00 - General
- Browse content in H1 - Structure and Scope of Government
- H10 - General
- H11 - Structure, Scope, and Performance of Government
- Browse content in H2 - Taxation, Subsidies, and Revenue
- H20 - General
- H21 - Efficiency; Optimal Taxation
- H22 - Incidence
- H23 - Externalities; Redistributive Effects; Environmental Taxes and Subsidies
- H24 - Personal Income and Other Nonbusiness Taxes and Subsidies; includes inheritance and gift taxes
- H25 - Business Taxes and Subsidies
- H26 - Tax Evasion and Avoidance
- Browse content in H3 - Fiscal Policies and Behavior of Economic Agents
- H31 - Household
- Browse content in H4 - Publicly Provided Goods
- H40 - General
- H41 - Public Goods
- H42 - Publicly Provided Private Goods
- H44 - Publicly Provided Goods: Mixed Markets
- Browse content in H5 - National Government Expenditures and Related Policies
- H50 - General
- H51 - Government Expenditures and Health
- H52 - Government Expenditures and Education
- H53 - Government Expenditures and Welfare Programs
- H54 - Infrastructures; Other Public Investment and Capital Stock
- H55 - Social Security and Public Pensions
- H56 - National Security and War
- H57 - Procurement
- Browse content in H6 - National Budget, Deficit, and Debt
- H63 - Debt; Debt Management; Sovereign Debt
- Browse content in H7 - State and Local Government; Intergovernmental Relations
- H70 - General
- H71 - State and Local Taxation, Subsidies, and Revenue
- H73 - Interjurisdictional Differentials and Their Effects
- H75 - State and Local Government: Health; Education; Welfare; Public Pensions
- H76 - State and Local Government: Other Expenditure Categories
- H77 - Intergovernmental Relations; Federalism; Secession
- Browse content in H8 - Miscellaneous Issues
- H81 - Governmental Loans; Loan Guarantees; Credits; Grants; Bailouts
- H83 - Public Administration; Public Sector Accounting and Audits
- H87 - International Fiscal Issues; International Public Goods
- Browse content in I - Health, Education, and Welfare
- Browse content in I0 - General
- I00 - General
- Browse content in I1 - Health
- I10 - General
- I11 - Analysis of Health Care Markets
- I12 - Health Behavior
- I13 - Health Insurance, Public and Private
- I14 - Health and Inequality
- I15 - Health and Economic Development
- I18 - Government Policy; Regulation; Public Health
- Browse content in I2 - Education and Research Institutions
- I20 - General
- I21 - Analysis of Education
- I22 - Educational Finance; Financial Aid
- I23 - Higher Education; Research Institutions
- I24 - Education and Inequality
- I25 - Education and Economic Development
- I26 - Returns to Education
- I28 - Government Policy
- Browse content in I3 - Welfare, Well-Being, and Poverty
- I30 - General
- I31 - General Welfare
- I32 - Measurement and Analysis of Poverty
- I38 - Government Policy; Provision and Effects of Welfare Programs
- Browse content in J - Labor and Demographic Economics
- Browse content in J0 - General
- J00 - General
- J01 - Labor Economics: General
- J08 - Labor Economics Policies
- Browse content in J1 - Demographic Economics
- J10 - General
- J11 - Demographic Trends, Macroeconomic Effects, and Forecasts
- J12 - Marriage; Marital Dissolution; Family Structure; Domestic Abuse
- J13 - Fertility; Family Planning; Child Care; Children; Youth
- J14 - Economics of the Elderly; Economics of the Handicapped; Non-Labor Market Discrimination
- J15 - Economics of Minorities, Races, Indigenous Peoples, and Immigrants; Non-labor Discrimination
- J16 - Economics of Gender; Non-labor Discrimination
- J18 - Public Policy
- Browse content in J2 - Demand and Supply of Labor
- J20 - General
- J21 - Labor Force and Employment, Size, and Structure
- J22 - Time Allocation and Labor Supply
- J23 - Labor Demand
- J24 - Human Capital; Skills; Occupational Choice; Labor Productivity
- J26 - Retirement; Retirement Policies
- Browse content in J3 - Wages, Compensation, and Labor Costs
- J30 - General
- J31 - Wage Level and Structure; Wage Differentials
- J33 - Compensation Packages; Payment Methods
- J38 - Public Policy
- Browse content in J4 - Particular Labor Markets
- J40 - General
- J42 - Monopsony; Segmented Labor Markets
- J44 - Professional Labor Markets; Occupational Licensing
- J45 - Public Sector Labor Markets
- J48 - Public Policy
- J49 - Other
- Browse content in J5 - Labor-Management Relations, Trade Unions, and Collective Bargaining
- J50 - General
- J51 - Trade Unions: Objectives, Structure, and Effects
- J53 - Labor-Management Relations; Industrial Jurisprudence
- Browse content in J6 - Mobility, Unemployment, Vacancies, and Immigrant Workers
- J60 - General
- J61 - Geographic Labor Mobility; Immigrant Workers
- J62 - Job, Occupational, and Intergenerational Mobility
- J63 - Turnover; Vacancies; Layoffs
- J64 - Unemployment: Models, Duration, Incidence, and Job Search
- J65 - Unemployment Insurance; Severance Pay; Plant Closings
- J68 - Public Policy
- Browse content in J7 - Labor Discrimination
- J71 - Discrimination
- J78 - Public Policy
- Browse content in J8 - Labor Standards: National and International
- J81 - Working Conditions
- J88 - Public Policy
- Browse content in K - Law and Economics
- Browse content in K0 - General
- K00 - General
- Browse content in K1 - Basic Areas of Law
- K14 - Criminal Law
- K2 - Regulation and Business Law
- Browse content in K3 - Other Substantive Areas of Law
- K31 - Labor Law
- K36 - Family and Personal Law
- Browse content in K4 - Legal Procedure, the Legal System, and Illegal Behavior
- K40 - General
- K41 - Litigation Process
- K42 - Illegal Behavior and the Enforcement of Law
- Browse content in L - Industrial Organization
- Browse content in L0 - General
- L00 - General
- Browse content in L1 - Market Structure, Firm Strategy, and Market Performance
- L10 - General
- L11 - Production, Pricing, and Market Structure; Size Distribution of Firms
- L13 - Oligopoly and Other Imperfect Markets
- L14 - Transactional Relationships; Contracts and Reputation; Networks
- L15 - Information and Product Quality; Standardization and Compatibility
- L16 - Industrial Organization and Macroeconomics: Industrial Structure and Structural Change; Industrial Price Indices
- L19 - Other
- Browse content in L2 - Firm Objectives, Organization, and Behavior
- L21 - Business Objectives of the Firm
- L22 - Firm Organization and Market Structure
- L23 - Organization of Production
- L24 - Contracting Out; Joint Ventures; Technology Licensing
- L25 - Firm Performance: Size, Diversification, and Scope
- L26 - Entrepreneurship
- Browse content in L3 - Nonprofit Organizations and Public Enterprise
- L33 - Comparison of Public and Private Enterprises and Nonprofit Institutions; Privatization; Contracting Out
- Browse content in L4 - Antitrust Issues and Policies
- L40 - General
- L41 - Monopolization; Horizontal Anticompetitive Practices
- L42 - Vertical Restraints; Resale Price Maintenance; Quantity Discounts
- Browse content in L5 - Regulation and Industrial Policy
- L50 - General
- L51 - Economics of Regulation
- Browse content in L6 - Industry Studies: Manufacturing
- L60 - General
- L62 - Automobiles; Other Transportation Equipment; Related Parts and Equipment
- L63 - Microelectronics; Computers; Communications Equipment
- L66 - Food; Beverages; Cosmetics; Tobacco; Wine and Spirits
- Browse content in L7 - Industry Studies: Primary Products and Construction
- L71 - Mining, Extraction, and Refining: Hydrocarbon Fuels
- L73 - Forest Products
- Browse content in L8 - Industry Studies: Services
- L81 - Retail and Wholesale Trade; e-Commerce
- L83 - Sports; Gambling; Recreation; Tourism
- L84 - Personal, Professional, and Business Services
- L86 - Information and Internet Services; Computer Software
- Browse content in L9 - Industry Studies: Transportation and Utilities
- L91 - Transportation: General
- L93 - Air Transportation
- L94 - Electric Utilities
- Browse content in M - Business Administration and Business Economics; Marketing; Accounting; Personnel Economics
- Browse content in M1 - Business Administration
- M11 - Production Management
- M12 - Personnel Management; Executives; Executive Compensation
- M14 - Corporate Culture; Social Responsibility
- Browse content in M2 - Business Economics
- M21 - Business Economics
- Browse content in M3 - Marketing and Advertising
- M31 - Marketing
- M37 - Advertising
- Browse content in M4 - Accounting and Auditing
- M42 - Auditing
- M48 - Government Policy and Regulation
- Browse content in M5 - Personnel Economics
- M50 - General
- M51 - Firm Employment Decisions; Promotions
- M52 - Compensation and Compensation Methods and Their Effects
- M53 - Training
- M54 - Labor Management
- Browse content in N - Economic History
- Browse content in N0 - General
- N00 - General
- N01 - Development of the Discipline: Historiographical; Sources and Methods
- Browse content in N1 - Macroeconomics and Monetary Economics; Industrial Structure; Growth; Fluctuations
- N10 - General, International, or Comparative
- N11 - U.S.; Canada: Pre-1913
- N12 - U.S.; Canada: 1913-
- N13 - Europe: Pre-1913
- N17 - Africa; Oceania
- Browse content in N2 - Financial Markets and Institutions
- N20 - General, International, or Comparative
- N22 - U.S.; Canada: 1913-
- N23 - Europe: Pre-1913
- Browse content in N3 - Labor and Consumers, Demography, Education, Health, Welfare, Income, Wealth, Religion, and Philanthropy
- N30 - General, International, or Comparative
- N31 - U.S.; Canada: Pre-1913
- N32 - U.S.; Canada: 1913-
- N33 - Europe: Pre-1913
- N34 - Europe: 1913-
- N36 - Latin America; Caribbean
- N37 - Africa; Oceania
- Browse content in N4 - Government, War, Law, International Relations, and Regulation
- N40 - General, International, or Comparative
- N41 - U.S.; Canada: Pre-1913
- N42 - U.S.; Canada: 1913-
- N43 - Europe: Pre-1913
- N44 - Europe: 1913-
- N45 - Asia including Middle East
- N47 - Africa; Oceania
- Browse content in N5 - Agriculture, Natural Resources, Environment, and Extractive Industries
- N50 - General, International, or Comparative
- N51 - U.S.; Canada: Pre-1913
- Browse content in N6 - Manufacturing and Construction
- N63 - Europe: Pre-1913
- Browse content in N7 - Transport, Trade, Energy, Technology, and Other Services
- N71 - U.S.; Canada: Pre-1913
- Browse content in N8 - Micro-Business History
- N82 - U.S.; Canada: 1913-
- Browse content in N9 - Regional and Urban History
- N91 - U.S.; Canada: Pre-1913
- N92 - U.S.; Canada: 1913-
- N93 - Europe: Pre-1913
- N94 - Europe: 1913-
- Browse content in O - Economic Development, Innovation, Technological Change, and Growth
- Browse content in O1 - Economic Development
- O10 - General
- O11 - Macroeconomic Analyses of Economic Development
- O12 - Microeconomic Analyses of Economic Development
- O13 - Agriculture; Natural Resources; Energy; Environment; Other Primary Products
- O14 - Industrialization; Manufacturing and Service Industries; Choice of Technology
- O15 - Human Resources; Human Development; Income Distribution; Migration
- O16 - Financial Markets; Saving and Capital Investment; Corporate Finance and Governance
- O17 - Formal and Informal Sectors; Shadow Economy; Institutional Arrangements
- O18 - Urban, Rural, Regional, and Transportation Analysis; Housing; Infrastructure
- O19 - International Linkages to Development; Role of International Organizations
- Browse content in O2 - Development Planning and Policy
- O23 - Fiscal and Monetary Policy in Development
- O25 - Industrial Policy
- Browse content in O3 - Innovation; Research and Development; Technological Change; Intellectual Property Rights
- O30 - General
- O31 - Innovation and Invention: Processes and Incentives
- O32 - Management of Technological Innovation and R&D
- O33 - Technological Change: Choices and Consequences; Diffusion Processes
- O34 - Intellectual Property and Intellectual Capital
- O38 - Government Policy
- Browse content in O4 - Economic Growth and Aggregate Productivity
- O40 - General
- O41 - One, Two, and Multisector Growth Models
- O43 - Institutions and Growth
- O44 - Environment and Growth
- O47 - Empirical Studies of Economic Growth; Aggregate Productivity; Cross-Country Output Convergence
- Browse content in O5 - Economywide Country Studies
- O52 - Europe
- O53 - Asia including Middle East
- O55 - Africa
- Browse content in P - Economic Systems
- Browse content in P0 - General
- P00 - General
- Browse content in P1 - Capitalist Systems
- P10 - General
- P16 - Political Economy
- P17 - Performance and Prospects
- P18 - Energy: Environment
- Browse content in P2 - Socialist Systems and Transitional Economies
- P26 - Political Economy; Property Rights
- Browse content in P3 - Socialist Institutions and Their Transitions
- P37 - Legal Institutions; Illegal Behavior
- Browse content in P4 - Other Economic Systems
- P48 - Political Economy; Legal Institutions; Property Rights; Natural Resources; Energy; Environment; Regional Studies
- Browse content in P5 - Comparative Economic Systems
- P51 - Comparative Analysis of Economic Systems
- Browse content in Q - Agricultural and Natural Resource Economics; Environmental and Ecological Economics
- Browse content in Q1 - Agriculture
- Q10 - General
- Q12 - Micro Analysis of Farm Firms, Farm Households, and Farm Input Markets
- Q13 - Agricultural Markets and Marketing; Cooperatives; Agribusiness
- Q14 - Agricultural Finance
- Q15 - Land Ownership and Tenure; Land Reform; Land Use; Irrigation; Agriculture and Environment
- Q16 - R&D; Agricultural Technology; Biofuels; Agricultural Extension Services
- Browse content in Q2 - Renewable Resources and Conservation
- Q25 - Water
- Browse content in Q3 - Nonrenewable Resources and Conservation
- Q32 - Exhaustible Resources and Economic Development
- Q34 - Natural Resources and Domestic and International Conflicts
- Browse content in Q4 - Energy
- Q41 - Demand and Supply; Prices
- Q48 - Government Policy
- Browse content in Q5 - Environmental Economics
- Q50 - General
- Q51 - Valuation of Environmental Effects
- Q53 - Air Pollution; Water Pollution; Noise; Hazardous Waste; Solid Waste; Recycling
- Q54 - Climate; Natural Disasters; Global Warming
- Q56 - Environment and Development; Environment and Trade; Sustainability; Environmental Accounts and Accounting; Environmental Equity; Population Growth
- Q58 - Government Policy
- Browse content in R - Urban, Rural, Regional, Real Estate, and Transportation Economics
- Browse content in R0 - General
- R00 - General
- Browse content in R1 - General Regional Economics
- R11 - Regional Economic Activity: Growth, Development, Environmental Issues, and Changes
- R12 - Size and Spatial Distributions of Regional Economic Activity
- R13 - General Equilibrium and Welfare Economic Analysis of Regional Economies
- Browse content in R2 - Household Analysis
- R20 - General
- R23 - Regional Migration; Regional Labor Markets; Population; Neighborhood Characteristics
- R28 - Government Policy
- Browse content in R3 - Real Estate Markets, Spatial Production Analysis, and Firm Location
- R30 - General
- R31 - Housing Supply and Markets
- R38 - Government Policy
- Browse content in R4 - Transportation Economics
- R40 - General
- R41 - Transportation: Demand, Supply, and Congestion; Travel Time; Safety and Accidents; Transportation Noise
- R48 - Government Pricing and Policy
- Browse content in Z - Other Special Topics
- Browse content in Z1 - Cultural Economics; Economic Sociology; Economic Anthropology
- Z10 - General
- Z12 - Religion
- Z13 - Economic Sociology; Economic Anthropology; Social and Economic Stratification
- Advance Articles
- Editor's Choice
- Author Guidelines
- Submission Site
- Open Access Options
- Self-Archiving Policy
- Why Submit?
- About The Quarterly Journal of Economics
- Editorial Board
- Advertising and Corporate Services
- Journals Career Network
- Dispatch Dates
- Journals on Oxford Academic
- Books on Oxford Academic

- < Previous
Machine Learning as a Tool for Hypothesis Generation
- Article contents
- Figures & tables
- Supplementary Data
Jens Ludwig, Sendhil Mullainathan, Machine Learning as a Tool for Hypothesis Generation, The Quarterly Journal of Economics , Volume 139, Issue 2, May 2024, Pages 751–827, https://doi.org/10.1093/qje/qjad055
- Permissions Icon Permissions
While hypothesis testing is a highly formalized activity, hypothesis generation remains largely informal. We propose a systematic procedure to generate novel hypotheses about human behavior, which uses the capacity of machine learning algorithms to notice patterns people might not. We illustrate the procedure with a concrete application: judge decisions about whom to jail. We begin with a striking fact: the defendant’s face alone matters greatly for the judge’s jailing decision. In fact, an algorithm given only the pixels in the defendant’s mug shot accounts for up to half of the predictable variation. We develop a procedure that allows human subjects to interact with this black-box algorithm to produce hypotheses about what in the face influences judge decisions. The procedure generates hypotheses that are both interpretable and novel: they are not explained by demographics (e.g., race) or existing psychology research, nor are they already known (even if tacitly) to people or experts. Though these results are specific, our procedure is general. It provides a way to produce novel, interpretable hypotheses from any high-dimensional data set (e.g., cell phones, satellites, online behavior, news headlines, corporate filings, and high-frequency time series). A central tenet of our article is that hypothesis generation is a valuable activity, and we hope this encourages future work in this largely “prescientific” stage of science.
Personal account
- Sign in with email/username & password
- Get email alerts
- Save searches
- Purchase content
- Activate your purchase/trial code
- Add your ORCID iD
Institutional access
Sign in with a library card.
- Sign in with username/password
- Recommend to your librarian
- Institutional account management
- Get help with access
Access to content on Oxford Academic is often provided through institutional subscriptions and purchases. If you are a member of an institution with an active account, you may be able to access content in one of the following ways:
IP based access
Typically, access is provided across an institutional network to a range of IP addresses. This authentication occurs automatically, and it is not possible to sign out of an IP authenticated account.
Choose this option to get remote access when outside your institution. Shibboleth/Open Athens technology is used to provide single sign-on between your institution’s website and Oxford Academic.
- Click Sign in through your institution.
- Select your institution from the list provided, which will take you to your institution's website to sign in.
- When on the institution site, please use the credentials provided by your institution. Do not use an Oxford Academic personal account.
- Following successful sign in, you will be returned to Oxford Academic.
If your institution is not listed or you cannot sign in to your institution’s website, please contact your librarian or administrator.
Enter your library card number to sign in. If you cannot sign in, please contact your librarian.
Society Members
Society member access to a journal is achieved in one of the following ways:
Sign in through society site
Many societies offer single sign-on between the society website and Oxford Academic. If you see ‘Sign in through society site’ in the sign in pane within a journal:
- Click Sign in through society site.
- When on the society site, please use the credentials provided by that society. Do not use an Oxford Academic personal account.
If you do not have a society account or have forgotten your username or password, please contact your society.
Sign in using a personal account
Some societies use Oxford Academic personal accounts to provide access to their members. See below.
A personal account can be used to get email alerts, save searches, purchase content, and activate subscriptions.
Some societies use Oxford Academic personal accounts to provide access to their members.
Viewing your signed in accounts
Click the account icon in the top right to:
- View your signed in personal account and access account management features.
- View the institutional accounts that are providing access.
Signed in but can't access content
Oxford Academic is home to a wide variety of products. The institutional subscription may not cover the content that you are trying to access. If you believe you should have access to that content, please contact your librarian.
For librarians and administrators, your personal account also provides access to institutional account management. Here you will find options to view and activate subscriptions, manage institutional settings and access options, access usage statistics, and more.
Short-term Access
To purchase short-term access, please sign in to your personal account above.
Don't already have a personal account? Register
| Month: | Total Views: |
|---|---|
| January 2024 | 927 |
| February 2024 | 626 |
| March 2024 | 509 |
| April 2024 | 1,754 |
| May 2024 | 896 |
| June 2024 | 618 |
| July 2024 | 386 |
| August 2024 | 75 |
Email alerts
Citing articles via.
- Recommend to Your Librarian
Affiliations
- Online ISSN 1531-4650
- Print ISSN 0033-5533
- Copyright © 2024 President and Fellows of Harvard College
- About Oxford Academic
- Publish journals with us
- University press partners
- What we publish
- New features
- Open access
- Rights and permissions
- Accessibility
- Advertising
- Media enquiries
- Oxford University Press
- Oxford Languages
- University of Oxford
Oxford University Press is a department of the University of Oxford. It furthers the University's objective of excellence in research, scholarship, and education by publishing worldwide
- Copyright © 2024 Oxford University Press
- Cookie settings
- Cookie policy
- Privacy policy
- Legal notice
This Feature Is Available To Subscribers Only
Sign In or Create an Account
This PDF is available to Subscribers Only
For full access to this pdf, sign in to an existing account, or purchase an annual subscription.
Machine Learning as a Tool for Hypothesis Generation
While hypothesis testing is a highly formalized activity, hypothesis generation remains largely informal. We propose a systematic procedure to generate novel hypotheses about human behavior, which uses the capacity of machine learning algorithms to notice patterns people might not. We illustrate the procedure with a concrete application: judge decisions about who to jail. We begin with a striking fact: The defendant’s face alone matters greatly for the judge’s jailing decision. In fact, an algorithm given only the pixels in the defendant’s mugshot accounts for up to half of the predictable variation. We develop a procedure that allows human subjects to interact with this black-box algorithm to produce hypotheses about what in the face influences judge decisions. The procedure generates hypotheses that are both interpretable and novel: They are not explained by demographics (e.g. race) or existing psychology research; nor are they already known (even if tacitly) to people or even experts. Though these results are specific, our procedure is general. It provides a way to produce novel, interpretable hypotheses from any high-dimensional dataset (e.g. cell phones, satellites, online behavior, news headlines, corporate filings, and high-frequency time series). A central tenet of our paper is that hypothesis generation is in and of itself a valuable activity, and hope this encourages future work in this largely “pre-scientific” stage of science.
This is a revised version of Chicago Booth working paper 22-15 “Algorithmic Behavioral Science: Machine Learning as a Tool for Scientific Discovery.” We gratefully acknowledge support from the Alfred P. Sloan Foundation, Emmanuel Roman, and the Center for Applied Artificial Intelligence at the University of Chicago. For valuable comments we thank Andrei Shliefer, Larry Katz and five anonymous referees, as well as Marianne Bertrand, Jesse Bruhn, Steven Durlauf, Joel Ferguson, Emma Harrington, Supreet Kaur, Matteo Magnaricotte, Dev Patel, Betsy Levy Paluck, Roberto Rocha, Evan Rose, Suproteem Sarkar, Josh Schwartzstein, Nick Swanson, Nadav Tadelis, Richard Thaler, Alex Todorov, Jenny Wang and Heather Yang, as well as seminar participants at Bocconi, Brown, Columbia, ETH Zurich, Harvard, MIT, Stanford, the University of California Berkeley, the University of Chicago, the University of Pennsylvania, the 2022 Behavioral Economics Annual Meetings and the 2022 NBER summer institute. For invaluable assistance with the data and analysis we thank Cecilia Cook, Logan Crowl, Arshia Elyaderani, and especially Jonas Knecht and James Ross. This research was reviewed by the University of Chicago Social and Behavioral Sciences Institutional Review Board (IRB20-0917) and deemed exempt because the project relies on secondary analysis of public data sources. All opinions and any errors are of course our own. The views expressed herein are those of the authors and do not necessarily reflect the views of the National Bureau of Economic Research.
MARC RIS BibTeΧ
Download Citation Data
Published Versions
Jens Ludwig & Sendhil Mullainathan, 2024. " Machine Learning as a Tool for Hypothesis Generation, " The Quarterly Journal of Economics, vol 139(2), pages 751-827.
Working Groups
Conferences, more from nber.
In addition to working papers , the NBER disseminates affiliates’ latest findings through a range of free periodicals — the NBER Reporter , the NBER Digest , the Bulletin on Retirement and Disability , the Bulletin on Health , and the Bulletin on Entrepreneurship — as well as online conference reports , video lectures , and interviews .

An official website of the United States government
The .gov means it’s official. Federal government websites often end in .gov or .mil. Before sharing sensitive information, make sure you’re on a federal government site.
The site is secure. The https:// ensures that you are connecting to the official website and that any information you provide is encrypted and transmitted securely.
- Publications
- Account settings
- My Bibliography
- Collections
- Citation manager
Save citation to file
Email citation, add to collections.
- Create a new collection
- Add to an existing collection
Add to My Bibliography
Your saved search, create a file for external citation management software, your rss feed.
- Search in PubMed
- Search in NLM Catalog
- Add to Search
Hypothesis generation, sparse categories, and the positive test strategy
Affiliation.
- 1 School of Psychology, University of Adelaide, South Australia 5005, Australia. [email protected]
- PMID: 21058871
- DOI: 10.1037/a0021110
We consider the situation in which a learner must induce the rule that explains an observed set of data but the hypothesis space of possible rules is not explicitly enumerated or identified. The first part of the article demonstrates that as long as hypotheses are sparse (i.e., index less than half of the possible entities in the domain) then a positive test strategy is near optimal. The second part of this article then demonstrates that a preference for sparse hypotheses (a sparsity bias) emerges as a natural consequence of the family resemblance principle; that is, it arises from the requirement that good rules index entities that are more similar to one another than they are to entities that do not satisfy the rule.
PubMed Disclaimer
Similar articles
- Learning overhypotheses with hierarchical Bayesian models. Kemp C, Perfors A, Tenenbaum JB. Kemp C, et al. Dev Sci. 2007 May;10(3):307-21. doi: 10.1111/j.1467-7687.2007.00585.x. Dev Sci. 2007. PMID: 17444972
- Effectiveness of Positive Hypothesis Testing in Inductive and Deductive Rule Learning. Laughlin PR, Bonner BL, Altermatt TW. Laughlin PR, et al. Organ Behav Hum Decis Process. 1999 Feb;77(2):130-146. doi: 10.1006/obhd.1998.2815. Organ Behav Hum Decis Process. 1999. PMID: 10069943
- Similarity, feature discovery, and the size principle. Navarro DJ, Perfors AF. Navarro DJ, et al. Acta Psychol (Amst). 2010 Mar;133(3):256-68. doi: 10.1016/j.actpsy.2009.10.008. Epub 2009 Dec 2. Acta Psychol (Amst). 2010. PMID: 19959157
- A Bayesian approach to generating tutorial hints in a collaborative medical problem-based learning system. Suebnukarn S, Haddawy P. Suebnukarn S, et al. Artif Intell Med. 2006 Sep;38(1):5-24. doi: 10.1016/j.artmed.2005.04.003. Epub 2005 Sep 23. Artif Intell Med. 2006. PMID: 16183267 Review.
- Bayesian ranking of sites for engineering safety improvements: decision parameter, treatability concept, statistical criterion, and spatial dependence. Miaou SP, Song JJ. Miaou SP, et al. Accid Anal Prev. 2005 Jul;37(4):699-720. doi: 10.1016/j.aap.2005.03.012. Epub 2005 Apr 12. Accid Anal Prev. 2005. PMID: 15949462 Review.
- Federated inference and belief sharing. Friston KJ, Parr T, Heins C, Constant A, Friedman D, Isomura T, Fields C, Verbelen T, Ramstead M, Clippinger J, Frith CD. Friston KJ, et al. Neurosci Biobehav Rev. 2024 Jan;156:105500. doi: 10.1016/j.neubiorev.2023.105500. Epub 2023 Dec 5. Neurosci Biobehav Rev. 2024. PMID: 38056542 Free PMC article. Review.
- Computational meaningfulness as the source of beneficial cognitive biases. Suomala J, Kauttonen J. Suomala J, et al. Front Psychol. 2023 May 2;14:1189704. doi: 10.3389/fpsyg.2023.1189704. eCollection 2023. Front Psychol. 2023. PMID: 37205079 Free PMC article.
- Structure learning principles of stereotype change. Gershman SJ, Cikara M. Gershman SJ, et al. Psychon Bull Rev. 2023 Aug;30(4):1273-1293. doi: 10.3758/s13423-023-02252-y. Epub 2023 Mar 27. Psychon Bull Rev. 2023. PMID: 36973602 Review.
- Adaptive search space pruning in complex strategic problems. Amir O, Tyomkin L, Hart Y. Amir O, et al. PLoS Comput Biol. 2022 Aug 10;18(8):e1010358. doi: 10.1371/journal.pcbi.1010358. eCollection 2022 Aug. PLoS Comput Biol. 2022. PMID: 35947588 Free PMC article.
- Social sampling and expressed attitudes: Authenticity preference and social extremeness aversion lead to social norm effects and polarization. Brown GDA, Lewandowsky S, Huang Z. Brown GDA, et al. Psychol Rev. 2022 Jan;129(1):18-48. doi: 10.1037/rev0000342. Psychol Rev. 2022. PMID: 35266789 Free PMC article.
Publication types
- Search in MeSH
LinkOut - more resources
Full text sources.
- American Psychological Association
- Ovid Technologies, Inc.
- Citation Manager
NCBI Literature Resources
MeSH PMC Bookshelf Disclaimer
The PubMed wordmark and PubMed logo are registered trademarks of the U.S. Department of Health and Human Services (HHS). Unauthorized use of these marks is strictly prohibited.
AI, Robot Neuroscientist: Reimagining Hypothesis Generation
Jiaqi shang , will xiao, send feedback.
Enter your feedback below and we'll get back to you as soon as possible. To submit a bug report or feature request, you can use the official OpenReview GitHub repository: Report an issue
BibTeX Record
- Data, AI, & Machine Learning
- Managing Technology
- Social Responsibility
- Workplace, Teams, & Culture
- AI & Machine Learning
- Diversity & Inclusion
- Big ideas Research Projects
- Artificial Intelligence and Business Strategy
- Responsible AI
- Future of the Workforce
- Future of Leadership
- All Research Projects
- AI in Action
- Most Popular
- The Truth Behind the Nursing Crisis
- Coaching for the Future-Forward Leader
- Measuring Culture

Our summer 2024 issue highlights ways to better support customers, partners, and employees, while our special report shows how organizations can advance their AI practice.
- Past Issues
- Upcoming Events
- Video Archive
- Me, Myself, and AI
- Three Big Points

Why Hypotheses Beat Goals

- Developing Strategy
- Skills & Learning

Not long ago, it became fashionable to embrace failure as a sign of a company’s willingness to take risks. This trend lost favor as executives recognized that what they wanted was learning, not necessarily failure. Every failure can be attributed to a raft of missteps, and many failures do not automatically contribute to future success.
Certainly, if companies want to aggressively pursue learning, they must accept that failures will happen. But the practice of simply setting goals and then being nonchalant if they fail is inadequate.
Instead, companies should focus organizational energy on hypothesis generation and testing. Hypotheses force individuals to articulate in advance why they believe a given course of action will succeed. A failure then exposes an incorrect hypothesis — which can more reliably convert into organizational learning.
What Exactly Is a Hypothesis?
When my son was in second grade, his teacher regularly introduced topics by asking students to state some initial assumptions. For example, she introduced a unit on whales by asking: How big is a blue whale? The students all knew blue whales were big, but how big? Guesses ranged from the size of the classroom to the size of two elephants to the length of all the students in class lined up in a row. Students then set out to measure the classroom and the length of the row they formed, and they looked up the size of an elephant. They compared their results with the measurements of the whale and learned how close their estimates were.
Note that in this example, there is much more going on than just learning the size of a whale. Students were learning to recognize assumptions, make intelligent guesses based on those assumptions, determine how to test the accuracy of their guesses, and then assess the results.
This is the essence of hypothesis generation. A hypothesis emerges from a set of underlying assumptions. It is an articulation of how those assumptions are expected to play out in a given context. In short, a hypothesis is an intelligent, articulated guess that is the basis for taking action and assessing outcomes.
Get Updates on Transformative Leadership
Evidence-based resources that can help you lead your team more effectively, delivered to your inbox monthly.
Please enter a valid email address
Thank you for signing up
Privacy Policy
Hypothesis generation in companies becomes powerful if people are forced to articulate and justify their assumptions. It makes the path from hypothesis to expected outcomes clear enough that, should the anticipated outcomes fail to materialize, people will agree that the hypothesis was faulty.
Building a culture of effective hypothesizing can lead to more thoughtful actions and a better understanding of outcomes. Not only will failures be more likely to lead to future successes, but successes will foster future successes.
Why Is Hypothesis Generation Important?
Digital technologies are creating new business opportunities, but as I’ve noted in earlier columns , companies must experiment to learn both what is possible and what customers want. Most companies are relying on empowered, agile teams to conduct these experiments. That’s because teams can rapidly hypothesize, test, and learn.
Hypothesis generation contrasts starkly with more traditional management approaches designed for process optimization. Process optimization involves telling employees both what to do and how to do it. Process optimization is fine for stable business processes that have been standardized for consistency. (Standardized processes can usually be automated, specifically because they are stable.) Increasingly, however, companies need their people to steer efforts that involve uncertainty and change. That’s when organizational learning and hypothesis generation are particularly important.
Shifting to a culture that encourages empowered teams to hypothesize isn’t easy. Established hierarchies have developed managers accustomed to directing employees on how to accomplish their objectives. Those managers invariably rose to power by being the smartest person in the room. Such managers can struggle with the requirements for leading empowered teams. They may recognize the need to hold teams accountable for outcomes rather than specific tasks, but they may not be clear about how to guide team efforts.
Some newer companies have baked this concept into their organizational structure. Leaders at the Swedish digital music service Spotify note that it is essential to provide clear missions to teams . A clear mission sets up a team to articulate measurable goals. Teams can then hypothesize how they can best accomplish those goals. The role of leaders is to quiz teams about their hypotheses and challenge their logic if those hypotheses appear to lack support.
A leader at another company told me that accountability for outcomes starts with hypotheses. If a team cannot articulate what it intends to do and what outcomes it anticipates, it is unlikely that team will deliver on its mission. In short, the success of empowered teams depends upon management shifting from directing employees to guiding their development of hypotheses. This is how leaders hold their teams accountable for outcomes.
Members of empowered teams are not the only people who need to hone their ability to hypothesize. Leaders in companies that want to seize digital opportunities are learning through their experiments which strategies hold real promise for future success. They must, in effect, hypothesize about what will make the company successful in a digital economy. If they take the next step and articulate those hypotheses and establish metrics for assessing the outcomes of their actions, they will facilitate learning about the company’s long-term success. Hypothesis generation can become a critical competency throughout a company.
How Does a Company Become Proficient at Hypothesizing?
Most business leaders have embraced the importance of evidence-based decision-making. But developing a culture of evidence-based decision-making by promoting hypothesis generation is a new challenge.
For one thing, many hypotheses are sloppy. While many people naturally hypothesize and take actions based on their hypotheses, their underlying assumptions may go unexamined. Often, they don’t clearly articulate the premise itself. The better hypotheses are straightforward and succinctly written. They’re pointed about the suppositions they’re based on. And they’re shared, allowing an audience to examine the assumptions (are they accurate?) and the postulate itself (is it an intelligent, articulated guess that is the basis for taking action and assessing outcomes?).
Related Articles
Seven-Eleven Japan offers a case in how do to hypotheses right.
For over 30 years, Seven-Eleven Japan was the most profitable retailer in Japan. It achieved that stature by relying on each store’s salesclerks to decide what items to stock on that store’s shelves. Many of the salesclerks were part-time, but they were each responsible for maximizing turnover for one part of the store’s inventory, and they received detailed reports so they could monitor their own performance.
The language of hypothesis formulation was part of their process. Each week, Seven-Eleven Japan counselors visited the stores and asked salesclerks three questions:
- What did you hypothesize this week? (That is, what did you order?)
- How did you do? (That is, did you sell what you ordered?)
- How will you do better next week? (That is, how will you incorporate the learning?)
By repeatedly asking these questions and checking the data for results, counselors helped people throughout the company hypothesize, test, and learn. The result was consistently strong inventory turnover and profitability.
How can other companies get started on this path? Evidence-based decision-making requires data — good data, as the Seven-Eleven Japan example shows. But rather than get bogged down with the limits of a company’s data, I would argue that companies can start to change their culture by constantly exposing individual hypotheses. Those hypotheses will highlight what data matters most — and the need of teams to test hypotheses will help generate enthusiasm for cleaning up bad data. A sense of accountability for generating and testing hypotheses then fosters a culture of evidence-based decision-making.
The uncertainties and speed of change in the current business environment render traditional management approaches ineffective. To create the agile, evidence-based, learning culture your business needs to succeed in a digital economy, I suggest that instead of asking What is your goal? you make it a habit to ask What is your hypothesis?
About the Author
Jeanne Ross is principal research scientist for MIT’s Center for Information Systems Research . Follow CISR on Twitter @mit_cisr .
More Like This
Add a comment cancel reply.
You must sign in to post a comment. First time here? Sign up for a free account : Comment on articles and get access to many more articles.
Comment (1)
Richard jones.
Building an Ethical and Trustworthy Biomedical AI Ecosystem for the Translational and Clinical Integration of Foundational Models
- Sankar Baradwaj, Simha
- Gilliland, Destiny
- Rincon, Jack
- Hermjakob, Henning
- Adam, Irsyad
- Lemaster, Gwyneth
- Watson, Karol
- Ping, Peipei
Foundational Models (FMs) are emerging as the cornerstone of the biomedical AI ecosystem due to their ability to represent and contextualize multimodal biomedical data. These capabilities allow FMs to be adapted for various tasks, including biomedical reasoning, hypothesis generation, and clinical decision-making. This review paper examines the foundational components of an ethical and trustworthy AI (ETAI) biomedical ecosystem centered on FMs, highlighting key challenges and solutions. The ETAI biomedical ecosystem is defined by seven key components which collectively integrate FMs into clinical settings: Data Lifecycle Management, Data Processing, Model Development, Model Evaluation, Clinical Translation, AI Governance and Regulation, and Stakeholder Engagement. While the potential of biomedical AI is immense, it requires heightened ethical vigilance and responsibility. For instance, biases can arise from data, algorithms, and user interactions, necessitating techniques to assess and mitigate bias prior to, during, and after model development. Moreover, interpretability, explainability, and accountability are key to ensuring the trustworthiness of AI systems, while workflow transparency in training, testing, and evaluation is crucial for reproducibility. Safeguarding patient privacy and security involves addressing challenges in data access, cloud data privacy, patient re-identification, membership inference attacks, and data memorization. Additionally, AI governance and regulation are essential for ethical AI use in biomedicine, guided by global standards. Furthermore, stakeholder engagement is essential at every stage of the AI pipeline and lifecycle for clinical translation. By adhering to these principles, we can harness the transformative potential of AI and develop an ETAI ecosystem.
- Computer Science - Computers and Society;
- Computer Science - Artificial Intelligence
Grab your spot at the free arXiv Accessibility Forum
Help | Advanced Search
Computer Science > Computation and Language
Title: automated review generation method based on large language models.
Abstract: Literature research, vital for scientific advancement, is overwhelmed by the vast ocean of available information. Addressing this, we propose an automated review generation method based on Large Language Models (LLMs) to streamline literature processing and reduce cognitive load. In case study on propane dehydrogenation (PDH) catalysts, our method swiftly generated comprehensive reviews from 343 articles, averaging seconds per article per LLM account. Extended analysis of 1041 articles provided deep insights into catalysts' composition, structure, and performance. Recognizing LLMs' hallucinations, we employed a multi-layered quality control strategy, ensuring our method's reliability and effective hallucination mitigation. Expert verification confirms the accuracy and citation integrity of generated reviews, demonstrating LLM hallucination risks reduced to below 0.5% with over 95% confidence. Released Windows application enables one-click review generation, aiding researchers in tracking advancements and recommending literature. This approach showcases LLMs' role in enhancing scientific research productivity and sets the stage for further exploration.
| Comments: | 16 pages, 3 figures, 3 tables |
| Subjects: | Computation and Language (cs.CL); Artificial Intelligence (cs.AI); Data Analysis, Statistics and Probability (physics.data-an) |
| Cite as: | [cs.CL] |
| (or [cs.CL] for this version) | |
| Focus to learn more arXiv-issued DOI via DataCite |
Submission history
Access paper:.
- HTML (experimental)
- Other Formats
References & Citations
- INSPIRE HEP
- Google Scholar
- Semantic Scholar
BibTeX formatted citation
Bibliographic and Citation Tools
Code, data and media associated with this article, recommenders and search tools.
- Institution
arXivLabs: experimental projects with community collaborators
arXivLabs is a framework that allows collaborators to develop and share new arXiv features directly on our website.
Both individuals and organizations that work with arXivLabs have embraced and accepted our values of openness, community, excellence, and user data privacy. arXiv is committed to these values and only works with partners that adhere to them.
Have an idea for a project that will add value for arXiv's community? Learn more about arXivLabs .
Physical Review A
Covering atomic, molecular, and optical physics and quantum science.
- Collections
- Editorial Team
Hilbert space fragmentation from lattice geometry
Pieter h. harkema, michael iversen, and anne e. b. nielsen, phys. rev. a 110 , 023301 – published 2 august 2024.
- No Citing Articles
- INTRODUCTION
- HILBERT SPACE FRAGMENTATION
- AUTOCORRELATION FUNCTION OF LOCAL…
- ACKNOWLEDGMENTS
The eigenstate thermalization hypothesis describes how isolated many-body quantum systems reach thermal equilibrium. However, quantum many-body scars and Hilbert space fragmentation violate this hypothesis and cause nonthermal behavior. We demonstrate that Hilbert space fragmentation may arise from lattice geometry in a spin- 1 2 model that conserves the number of domain walls. We generalize a known, one-dimensional, scarred model to larger dimensions and show that this model displays Hilbert space fragmentation on the Vicsek fractal lattice and the two-dimensional lattice. Using Monte Carlo methods, the model is characterized as strongly fragmented on the Vicsek fractal lattice when the number of domain walls is either small or close to the maximal value. On the two-dimensional lattice, the model is strongly fragmented when the density of domain walls is low and weakly fragmented when the density of domain walls is high. Furthermore, we show that the fragmentation persists at a finite density of domain walls in the thermodynamic limit for the Vicsek fractal lattice and the two-dimensional lattice. We also show that the model displays signatures similar to Hilbert space fragmentation on a section of the second-generation hexaflake fractal lattice and a modified two-dimensional lattice. We study the autocorrelation function of local observables and demonstrate that the model displays nonthermal dynamics.
- Received 12 April 2024
- Accepted 16 July 2024
DOI: https://doi.org/10.1103/PhysRevA.110.023301
©2024 American Physical Society
Physics Subject Headings (PhySH)
- Research Areas
- Physical Systems
Authors & Affiliations
- Department of Physics and Astronomy, Aarhus University , DK-8000 Aarhus C, Denmark
- * These authors contributed equally to this work.
Article Text (Subscription Required)
References (subscription required).
Vol. 110, Iss. 2 — August 2024
Access Options
- Buy Article »
- Log in with individual APS Journal Account »
- Log in with a username/password provided by your institution »
- Get access through a U.S. public or high school library »

Authorization Required
Other options.
- Buy Article »
- Find an Institution with the Article »
Download & Share
Illustration of the considered lattices. Black dots show the lattice sites and red lines display nearest-neighbor edges. (a) The first-generation Vicsek fractal lattice. (b) The second-generation Vicsek fractal lattice is obtained from generation one by substituting all lattice sites with five new sites. (c) Similarly, the third-generation Vicsek fractal lattice is obtained from generation two by substituting all sites with generation one Vicsek fractal lattices. (d) The second-generation hexaflake fractal lattice. We consider the lattice consisting of three connected first-generation hexaflake fractal lattices (above the dashed line). (e) The two-dimensional lattice of size L x × L y = 5 × 4 . (f) The modified two-dimensional lattice constructed from four connected first-generation Vicsek fractal lattices.
The second-generation Vicsek fractal lattice padded with dynamically inactive sites along the boundary (dark red balls with black outlines and downward-pointing arrows). The figure illustrates a product state in the symmetry sector with n dw = 8 domain walls. Four sites are spin up (blue balls with upward pointing arrows) and the remaining sites are spin down (red balls). The sites inside the green box are denoted as an “active arm.” These sites may flip their spin and the domain walls may move around within the sublattice. Note, however, that the spin-down site connected to the active arm from the left (red ball at the center of the lattice) can not change its state to spin up because three or more of its nearest neighbors are always spin down. Therefore, this site effectively locks the dynamics within the active arm and all spin-down sites outside the green box can not be flipped. Similarly, the spin-up site inside the blue box is dynamically inactive and can not be flipped. Product states where all spins are dynamically inactive are denoted as “frozen states,” and we construct an exponential number of such states in Appendix pp2 .
The Hamiltonian in each symmetry sector for (a) the second-generation Vicsek fractal lattice illustrated in Fig. 1 with 25 dynamically active sites, (b) the section of the second-generation hexaflake fractal lattice illustrated in Fig. 1 with 21 dynamically active sites, (c) the two-dimensional lattice illustrated in Fig. 1 of size L x × L y = 5 × 4 , and (d) the modified two-dimensional lattice illustrated in Fig. 1 with 20 dynamically active sites. Gray pixels represent nonzero matrix elements and white pixels correspond to vanishing matrix elements. The figure illustrates the block-diagonal structure of the Hamiltonian operator due to Hilbert space fragmentation. n dw denotes the number of domain walls characterizing the symmetry sector, and D ≡ D n dw denotes the dimension of the symmetry sector. For the Vicsek fractal lattice, the Hamiltonian is only depicted up to n dw = 18 domain walls since the Krylov subspaces in the symmetry sector with n dw domain walls have the same sizes as the Krylov subspaces in the symmetry sector with n dw max − n dw domain walls as discussed in Appendix pp1 . The red asterisks mark symmetry sectors containing just a single Krylov subspace that spans the full sector.
The ratio between the dimension of the largest Krylov subspace d n dw max and the dimension of the symmetry sector D n dw as a function of the generation g of the Vicsek fractal lattice. Each graph corresponds to a fixed number of domain walls n dw . The data for generation g = 1 , 2 is exact while the data for generation g = 3 , 4 is obtained using Monte Carlo importance sampling as described in Appendix pp3 . The symmetry sector with n dw = 0 domain walls is one dimensional for all generations, and the ratio is unity. For all other considered sectors, the ratio decreases with increasing generation indicating that the system is strongly fragmented.
(a) The number of Krylov subspaces as a function of system size L x × L y . We vary the length of one side of the lattice L x while keeping the other side fixed at L y = 2 (dark blue dots), L y = 3 (light blue triangles), and L y = 4 (light red diamonds). We also increase the length L of both sides simultaneously L x × L y = L × L (dark red crosses). Notice that some data points are included in more than one of these groups and have multiple markers, e.g., L x × L y = 2 × 2 . The number of Krylov subspaces scales exponentially with system size and is larger than the lower bound (dashed line) obtained in Appendix pp4 . (b)–(d) The ratio between the dimension of the largest Krylov subspace and the dimension of the corresponding symmetry sector as a function of system size for various densities of domain walls. We consider (b) one-quarter filling n dw = n dw max / 4 , (c) half-filling n dw = n dw max / 2 , and (d) three-quarter filling n dw = 3 n dw max / 4 . The results indicate that the model is strongly fragmented for a low density of domain walls and weakly fragmented for a high density of domain walls.
The time average of the autocorrelation function for the four considered lattices and two symmetry sectors on the one-dimensional lattice of N sites. We consider the symmetry sector n dw = 6 ( D n dw = 5472 ) for the second-generation Vicsek fractal lattice, n dw = 14 ( D n dw = 7680 ) for the section of the second-generation hexaflake fractal lattice, n dw = 14 ( D n dw = 5323 ) for the two-dimensional lattice, and n dw = 6 ( D n dw = 3492 ) for the modified two-dimensional lattice. We consider the autocorrelation function of the operator s ̃ r z = s r z − 〈 s r z 〉 for the site r illustrated in the inset (inside red circle). We find similar results for other sites. In all cases, we consider parameters λ = J = 1 and Δ = 0.1 . The horizontal lines show the Mazur bound obtained from the projection operators onto the Krylov subspaces.
A part of the Vicsek fractal lattice of generation g = 3 . We consider the lattice to consist of generation-one fractal lattices. Some generation-one fractals are nearest neighbors with four other generation-one fractals (gray sites). We choose all sites to be spin down in these generation-one fractals. For generation-one fractals with two nearest neighbors, the spins are chosen among the three frozen configurations shown in the figure. For generation-one fractals with one nearest neighbor, we choose the spins among the four frozen configurations shown in the figure. We construct an exponential number of eigenstates of the Hamiltonian as the tensor product of these frozen configurations on the generation-one fractals.
The two-dimensional lattice L of size L x × L y (light red and blue circles). The lattice is padded with extra sites along its boundary (dark red circles) to ensure all sites in the original lattice have an even number of nearest neighbors. The extra sites are dynamically inactive and we choose them to be spin down. The lattice L does not contain the dynamically inactive sites. The sublattice L ′ ⊂ L (blue circles) form a 5-periodic pattern on L in the x and y directions. The lattice L is separated into four parts L A , L B , L C , and L D (gray boxes). Part L A is a rectangular lattice where both sides are a multiple of 5. Part L B contains no sites when mod 5 ( L x ) = 0 and L C contains no sites when mod 5 ( L y ) = 0 . When parts L B and L C are nonempty, they are rectangular lattices where one side is a multiple of 5. Part L D is empty when mod 5 ( L x ) = 0 or mod 5 ( L y ) = 0 . When part L D is not empty it is a rectangular lattice where neither side is a multiple of 5.
The time-averaged autocorrelation function on the Vicsek fractal lattice for the perturbed Hamiltonian from Eq. ( E1 ). The size of the perturbation is characterized by the ratio between the Frobenius norm of the perturbation matrix | H GOE | to that of the nonperturbed Hamiltonian | H | . The dashed line displays the Mazur bound obtained from the projection operators onto the Krylov subspaces.
Sign up to receive regular email alerts from Physical Review A
- Forgot your username/password?
- Create an account
Article Lookup
Paste a citation or doi, enter a citation.
An official website of the United States government
The .gov means it’s official. Federal government websites often end in .gov or .mil. Before sharing sensitive information, make sure you’re on a federal government site.
The site is secure. The https:// ensures that you are connecting to the official website and that any information you provide is encrypted and transmitted securely.
- Publications
- Account settings
Preview improvements coming to the PMC website in October 2024. Learn More or Try it out now .
- Advanced Search
- Journal List
- Epidemiol Infect
- v.147; 2019

Methods for generating hypotheses in human enteric illness outbreak investigations: a scoping review of the evidence
1 School of Public Health, University of Alberta, Edmonton, Canada
2 Outbreak Management Division, Centre for Food-borne, Environmental and Zoonotic Infectious Diseases, Public Health Agency of Canada, Guelph, Canada
3 Public Health Risk Sciences Division, National Microbiology Laboratory at Guelph, Public Health Agency of Canada, Guelph, Canada
M. Mascarenhas
Associated data.
For supplementary material accompanying this paper visit https://doi.org/10.1017/S0950268819001699.
Enteric illness outbreaks are complex events, therefore, outbreak investigators use many different hypothesis generation methods depending on the situation. This scoping review was conducted to describe methods used to generate a hypothesis during enteric illness outbreak investigations. The search included five databases and grey literature for articles published between 1 January 2000 and 2 May 2015. Relevance screening and article characterisation were conducted by two independent reviewers using pretested forms. There were 903 outbreaks that described hypothesis generation methods and 33 papers which focused on the evaluation of hypothesis generation methods. Common hypothesis generation methods described are analytic studies (64.8%), descriptive epidemiology (33.7%), food or environmental sampling (32.8%) and facility inspections (27.9%). The least common methods included the use of a single interviewer (0.4%) and investigation of outliers (0.4%). Most studies reported using two or more methods to generate hypotheses (81.2%), with 29.2% of studies reporting using four or more. The use of multiple different hypothesis generation methods both within and between outbreaks highlights the complexity of enteric illness outbreak investigations. Future research should examine the effectiveness of each method and the contexts for which each is most effective in efficiently leading to source identification.
Introduction
Enteric illnesses cause considerable morbidity and mortality worldwide. Waterborne enteric diseases cause 2 million deaths each year, the majority of which occur in children aged 5 and under [ 1 ]. Foodborne enteric diseases are responsible for 600 million illnesses and 420 000 deaths annually [ 2 ]. These illnesses impact the quality of life of those affected and result in enormous financial consequences for individuals and nations [ 3 ]. Although most enteric illnesses are transient, significant chronic sequelae associated with some foodborne pathogens can have long-term public health impacts [ 4 – 6 ].
Enteric illness outbreak investigations seek to identify the source of illnesses to prevent further illness in the population. Timely source identification is a key step towards reducing the incidence of enteric illness worldwide and can lead to change in public health policy or recommendations to prevent future outbreaks, such as changes to food manufacturing processes or regulations. Timely source identification can also lead to public health notices and recalls that may prevent further illnesses in a specific outbreak. Accurate source identification can also provide opportunities to learn more about known and emerging diseases, increase understanding of the impact of current disease prevention practices and improve public confidence in public health agencies responsiveness to disease outbreaks [ 7 ].
Outbreak investigations take many forms, depending on the pathogen, context, affected population and suspected route of transmission. Initial cases often alert public health officials that a possible outbreak is occurring. Once an outbreak has been identified a case definition is established to support case finding activities. As cases are identified, information is gathered about the outbreak to generate hypotheses about the potential source(s) and route(s) of exposure. Information can come from a range of sources, including the cases themselves, their friends or family, staff members of businesses and institutions, experts or literature and physical and environmental sampling and inspections. Taken together, this information supports the development of hypotheses about the source of the outbreak.
Hypothesis generation about both the potential source(s) and route(s) of exposure is a key step in outbreak investigations, as it begins the process of narrowing the search for the transmission vehicle. Although some hypothesis generation methods have been described in summaries of outbreak investigation steps [ 7 , 8 ], the full range of possible methods used in outbreak investigations or the frequency that they are used is not readily available. We conducted a scoping review to summarise the methods for hypothesis generation used during human enteric illness investigations and to understand the frequency and breadth of methods, as well as to identify knowledge gaps and areas for future research.
A scoping review protocol was created a priori using the framework established by Arksey and O'Malley [ 9 ]. A copy of the protocol, including the search strategy, the screening tool and the data characterisation tool can be found in Supplementary Material S1. A full list of the articles identified in this scoping review can be found in Supplementary Material S2. A review team was established and included expertise in synthesis research, food safety, epidemiology and outbreak investigation.
The research question:
What methods have been used, or could be used, in human enteric illness outbreak investigations for hypothesis generation?
Search terms and strategy
A search algorithm (Supplementary Material S1) was constructed using key terms from 30 pre-selected relevant articles and implemented in five databases (PubMed, Scopus, Embase, Cumulative Index to Nursing and Allied Health Literature (CINAHL) and ProQuest Public Health) on 25 May 2015 with a date filter of 1 January 2000–25 May 2015.
The search was evaluated for capture sensitivity by searching reference lists of 12 randomly selected relevant primary methodology papers and 10 of the most recent relevant literature reviews in PubMed (Supplementary Material S1). The grey literature search targeted websites of government and research organisations, and relevant articles from Conference Proceedings (Supplementary Material S1). A total of 202 articles were identified by the grey literature search that were not captured by the search strategy and were added to the literature review ( Fig. 1 ). All citations were exported and de-duplicated in RefWorks (ProQuest, LLC), an online bibliographic management program, before being uploaded into a web-based systematic review management program, DistillerSR™ (Evidence Partners, Ottawa, Canada), for evaluation and characterisation.

PRISMA flow chart documenting the literature retrieval and inclusion/exclusion criteria for citations to identify methods of hypothesis generation during human illness investigations.
Relevance screening of abstracts and full-text citation
Each title and abstract was screened by two independent reviewers using a relevance screening form (Supplementary Material S1). Articles were included if they met the following criteria: (1) used or described methods applicable to enteric illness outbreak investigations to assist in hypothesis generation and source identification; (2) published after 1 January 2000 and (3) were reported in either English or French language. No geographic location was used as an exclusion criterion. The relevance screening form was pretested on 50 citations and resulted in a kappa agreement >0.8, indicating good agreement. Two reviewers screened each citation independently and conflicts were resolved by consensus.
Potentially relevant articles were procured, confirmed to be in English or French and relevant before broadly being characterised by two independent reviewers using a secondary relevance screening tool (Supplementary Material S1) to gather information on the outbreak, such as geographic location, type of pathogen, setting (single or general) and implicated source (Supplementary Material S1). This form was pretested on 10 papers to ensure good agreement and clarity within the form.
Data extraction and analysis
The data characterisation and utility tool was used to gather data on the hypothesis generation methods used in the outbreak investigation. The form contained check boxes for 23 known hypothesis generation methods and an option for reviewers to add other methods not captured in the form. Clearly established definitions were used to help data extractors distinguish between instances when a method was used for hypothesis generation or hypothesis testing. Hypothesis generation was defined as the process of developing one or more tentative explanations about the source of the outbreak used to inform further investigation. This was distinguished from hypothesis testing, which was defined as the process of confirming that a specific exposure is or is not the cause of an outbreak. Hypothesis testing is performed on a small number of suspect exposures and may include statistical testing or traceback investigation. Sometimes, when the hypothesis is refuted, additional rounds of hypothesis generation may be initiated. Several methods included in the form could be used for either hypothesis generation or for hypothesis testing in outbreak investigations. For example, analytic studies can be used to examine a wide range of exposures to help generate hypotheses about plausible sources. However, analytic studies can also be used to test a hypothesis when a specific source is suspected. Instances where methods were used to test a hypothesis were not relevant to this review and were not captured on the form. Where more than one outbreak was described in a single paper, multiple forms were completed to capture methods used in different investigations. This form was pretested on five papers to ensure agreement between reviewers was adequate and to improve the clarity of the questions/answers where necessary. Two reviewers independently reviewed each paper and disagreements between reviewers were discussed until a consensus was reached or settled with a third reviewer. Articles with no hypothesis generation methods described or with a known source at the outset of the investigation were excluded at this stage. Papers describing methodology, but not specific outbreak investigations, were identified and are described separately. Descriptive statistics were used to summarise the dataset using Stata 15 (StataCorp, 2017).
In total, there were 10 615 unique citations captured by the search ( Fig. 1 ). Of these, 889 (8.4%) papers were fully characterised and included 903 reported outbreaks (Supplementary Material S2). Of the reported outbreaks, 25 (2.8%) were described in 11 multi-outbreak articles and the remaining 878 (97.2%) were described in single outbreak articles ( Fig. 1 ).
The pathogens associated with the outbreaks included: bacteria ( n = 622, 68.9%), viruses ( n = 192, 21.3%), parasites ( n = 64, 7.1%), bio-toxins ( n = 3, 0.3%), fungi ( n = 1, 0.1%) and multiple pathogens ( n = 11, 1.2%). The pathogen was not identified in 10 (1.1%) outbreaks. In terms of outbreak source, 552 (61.1%) identified food as the source, while 103 (11.4%) identified water, 34 (3.8%) identified direct contact with animals, 25 (2.8%) identified person-to-person transmission, 25 (2.8%) identified multiple modes of transmission, 20 (2.2%) identified food-handlers, 8 (0.9%) identified soil or environment and 5 (0.6%) reported other modes of transmission as the source. In 131 (14.5%) of the outbreaks, no source was identified.
Hypothesis generation methods used in the enteric illness outbreak investigations are listed and defined in Table 1 . The majority ( n = 733, 81.2%) of investigations employed two or more methods to generate hypotheses; the median number of methods used was three (interquartile range: 2–4). Analytic studies ( n = 585, 64.8%) were the most commonly reported method category, followed by descriptive epidemiology ( n = 304, 33.7%), and food or environmental sampling ( n = 296, 32.8%). Uncommon methods included tracer testing ( n = 1, 0.1%), anthropological investigation ( n = 1, 0.1%) and industry consultation ( n = 1, 0.1%).
Description and frequency of methods used to generate a hypothesis in 903 human enteric illness outbreak investigations identified in scoping review citations
| Hypothesis generation method | Definition | (%) | Setting used |
|---|---|---|---|
| Questionnaires | |||
| Hypothesis generation questionnaire | Questionnaires designed to capture a large number of exposures to generate hypotheses about possible sources of infection; questions often related to food and water consumption, behavioural habits, travel activities and animal exposures; sometimes referred to as trawling or shot-gun questionnaires. | 182 (20.2) | Single: 41 General: 141 |
| Focused questionnaire | Developed for a specific outbreak investigation, often with a shorter, more focused list of exposures than hypothesis-generating questionnaire; types included questionnaires developed based on a specific menu, and questionnaires developed after initial, longer, questionnaires ruled out potential sources. | 159 (17.6) | Single: 96 General: 63 |
| Routine questionnaire | Administered as part of initial case (routine) follow-up, often prior to an outbreak being identified or laboratory testing for the pathogen; the questionnaires are usually brief, containing only common risk factors. | 133 (14.7) | Single: 43 General: 90 |
| Enhanced surveillance questionnaire | Standardised questionnaire routinely administered as part of an enhanced surveillance initiative for a specific pathogen. Administered to cases following laboratory confirmation for specific pathogens. | 13 (1.4) | Single: 3 General: 10 |
| Questionnaire, unspecified | Questionnaires used to identify exposures not described as either focused, routine, enhanced surveillance or hypothesis generating. | 127 (14.1) | Single: 42 General: 85 |
| Interviews & focus groups | |||
| In-person interviewing | Face-to-face interviews, sometimes in the cases' home | 50 (5.5) | Single: 27 General: 23 |
| Open-ended interviewing | Unstructured, exploratory interview with open-ended questions to collect a detailed exposure history. Questions included food preferences, routines, habits and usual activities. | 38 (4.2) | Single: 13 General: 25 |
| Iterative interviewing | Questionnaire items were modified as new cases were interviewed, based on additional information provided by new cases. Exposures reported were amended in the questionnaire for future cases. Previous cases may be re-interviewed with new questions. | 16 (1.8) | Single: 4 General: 12 |
| Centralised interviewing | All interviews were conducted by one organisation, with one or more interviewers. Close proximity of interviewers enabled regular discussion of common exposures, which were used to generate hypotheses. | 8 (0.9) | Single: 2 General: 6 |
| Focus groups | Multiple interviewers or cases were brought together to discuss exposures to identify commonalities. Group discussion prompted recall of previously forgotten exposures and improved investigators' understanding of plausible sources and transmission routes to support hypothesis generation. | 6 (0.7) | Single: 1 General: 5 |
| Single interviewer | All interviews were conducted by the same person, which facilitated hypothesis generation because one can more easily identify commonalities across cases during interviews. | 4 (0.4) | Single: 0 General: 4 |
| Food displays | Photographs or physical plates of food used during case interviews to help trigger better recall of exposures from cases. | 2 (0.2) | Single: 2 General: 0 |
| Industry consultation | Consultations with independent industry experts to help generate hypotheses about suspected food items of interest or sources of contamination in the food production process. | 1 (0.1) | Single: 0 General: 1 |
| Analytic methods | |||
| Analytic Study | An analytic study conducted in the absence of a clearly stated hypothesis. Used to identify significantly different exposures between cases and controls. Types included: case-control, cohort, case-cohort, case-chaos, case-case and case-crossover. | 585 (64.8) | Single: 345 General: 240 |
| Interesting descriptive epidemiology | Examination of unique or interesting features of person, place, or time to identify patterns that provided clues about potential sources of the outbreak. | 304 (33.7) | Single: 118 General: 186 |
| Investigation of sub-clusters | Investigation of a localised event or non-household setting, such as a restaurant, linked to two or more cases in the outbreak to help identify common exposures. | 37 (4.1) | Single: 4 General: 33 |
| Binomial Probability/comparison to population estimates | Case exposure frequencies were compared to background rates or population exposure estimates, often using binomial probability calculations, to generate hypotheses about likely sources. Hypotheses were based on a significantly higher level of exposure among cases compared to the baseline population data. | 30 (3.3) | Single: 0 General: 30 |
| Investigation of outliers | Examination of one or a subset of cases with unusual exposures or specific food preferences that differed from overall sample. This helped generate new hypotheses or narrow down the number of hypotheses. | 4 (0.4) | Single: 2 General: 2 |
| Sampling & inspection | |||
| Food or environmental sampling | Sampling available food items in homes or restaurants, or obtaining environmental swabs of food preparation areas or other plausible sources to identify, through laboratory testing, a source linked to the outbreak. | 296 (32.8) | Single: 202 General: 94 |
| Facility inspections | Inspection of a facility to identify possible sources of contamination and foods that might be implicated by such contamination; could involve inspecting food handling and storage practices, food preparation activities, employee hygiene, water sanitation systems, or reviewing policies and procedures. | 252 (27.9) | Single: 209 General: 43 |
| Food handler testing | Biological sampling of food handlers working at suspected food establishments. Used to identify, through laboratory testing, a source linked to the outbreak. | 23 (2.5) | Single: 19 General: 4 |
| Household inspection | Inspection of a case's home to identify possible sources of contamination and foods that might be implicated by such contamination. Could involve inventories of pantry items for comparison across cases to aid in hypothesis generation of common exposures. | 2 (0.2) | Single: 0 General: 2 |
| Other methods | |||
| Review of existing information | Reviewing information sources to generate hypotheses about previously reported exposures to the pathogen or biologically plausible exposures; sources included peer-reviewed scientific or grey literature, published reports, or disease surveillance systems. | 86 (9.5) | Single: 14 General: 72 |
| Epidemiology traceback | Traceback to determine whether food consumed by multiple cases commonly converges in the supply chain or to compare the distribution of illnesses to the distribution of a food commodity to see if patterns emerged to help generate hypotheses. | 56 (6.2) | Single: 10 General: 46 |
| Menu or recipe analysis | Review of a menu or recipes to verify exposures reported by cases, or to identify specific ingredients within reported meals. | 51 (5.7) | Single: 34 General: 17 |
| Purchase records | Records of sales transactions, such as receipts, bank statements, or loyalty card history, used to verify exposure, identify commonalities between cases, or obtain product details. Institutional purchase and delivery records reviewed to generate hypotheses about plausible outbreak sources. | 39 (4.3) | Single: 8 General: 31 |
| Anecdotal reports | Unverified reports or suspicions from cases/external sources, such as the public or medical professionals, about the potential source(s) of an outbreak. Obtained directly from individuals, or through online media such as web forums or social media. | 37 (4.1) | Single: 24 General: 13 |
| Spatial epidemiology | Spot-mapping or geo-mapping cases to identify potential location-based linkages across cases, such as common grocery stores, activities or neighbourhoods. | 6 (0.7) | Single: 2 General: 4 |
| Contact tracing/social network analysis | Identification of all people who came into contact with a case to provide clues regarding plausible sources of illness. | 3 (0.3) | Single: 2 General: 1 |
| Anthropological investigation | Team of anthropologists employing ethnographic techniques to understand culturally-specific exposures; helped develop culturally-appropriate questionnaire for hypothesis generation within local language and customs. | 1 (0.1) | Single: 0 General: 1 |
| Tracer testing | Fluorescent dyes placed in a water or sanitation system to understand connections and travel time of water or effluent, which helped generate hypotheses about sources of water contamination. | 1 (0.1) | Single: 1 General: 0 |
Single setting outbreaks
The proportion that each method was used within single setting outbreaks, such as a restaurant, nursing home, or event, is reported in Figure 2 . The most commonly reported methods used in single setting outbreaks included analytic studies ( n = 345, 27.2%), facility inspections ( n = 209, 16.5%) and food or environmental sampling ( n = 202, 15.9%). The least common methods used in single setting outbreaks included focus groups ( n = 1, 0.1%) and tracer testing ( n = 1, 0.1%). Binomial probability/comparison to population estimates, single interviewer and anthropological investigation were not reported in single setting outbreaks.

Hypothesis generation methods used in single setting outbreaks.
General population outbreaks
The proportion that each method was used in general population outbreaks, outbreaks not related to a single event or venue, is reported in Figure 3 . The most commonly used methods in general population outbreaks included analytic studies ( n = 240, 18.7%), interesting descriptive epidemiology ( n = 186, 14.5%) and hypothesis generation questionnaires ( n = 141, 11.0%). The least common methods used in general population outbreaks included anthropological investigation ( n = 1, 0.1%), contact tracing/social network analysis ( n = 1, 0.1%) and industry consultation ( n = 1, 0.1%). Tracer testing and food displays were not reported in general population outbreaks.

Hypothesis generation methods used in general population outbreaks.
Hypothesis generation innovation and trends 2000–2015
Trends in method use over the 15-year span were examined in 5-year increments (Supplementary Material S3). Small increases were observed in the use of anecdotal reports, purchase records, binomial probability/population comparison, facility inspections and review of existing information. A decline was observed in the use of analytic studies. Other methods had variable use over the time period or were relatively stable.
Methodology papers
Of the 10 615 citations screened, 33 (0.3%) methods papers were identified (Supplementary Material S2). These papers focused on evaluating existing methods or comparing standard vs. a novel approach to hypothesis generation (Supplementary Material S4). Of these, the most commonly discussed method was analytic studies ( n = 11, 33.3%). This included five on the validity of case-chaos methodology [ 10 – 14 ], two on case-case methodology [ 15 , 16 ], two on case-control methodology [ 17 , 18 ], one discussing the validity of case-cohort methodology [ 19 ] and one discussing the validity of case-crossover methodology [ 20 ].
The use of laboratory methods, including whole genome sequencing, was described in five (15.2%) papers [ 21 – 25 ]. Traceback procedures were explored in five (15.2%) papers, including three on the use of network analysis [ 26 – 28 ], one on the use of food flow information [ 29 ] and one examining the use of relational systems to identify sources common to different cases [ 30 ]. Four (12.1%) papers described broad outbreak investigation activities, which included the hypothesis generation step, one from the United Kingdom [ 31 ], one from Quebec, Canada [ 32 ], one from Minnesota [ 33 ] and one from the Centers for Disease Control and Prevention (CDC) in the United States [ 34 ]. Three (9.1%) papers explored interviewing techniques, two examining the use of computer assisted telephone interviews (CATI) technology [ 35 , 36 ] and one on when to collect interview-intensive dose-response data [ 37 ]. Three (9.1%) papers compared online questionnaires to phone or paper questionnaires [ 38 – 40 ]. Finally, one (3.0%) paper examined the use of mathematical topology methods to generate hypotheses [ 41 ] and another (3.0%) paper examined the use of sales record data to generate hypotheses [ 42 ].
The most commonly reported hypothesis generation methods identified in this scoping review included analytic studies, descriptive epidemiology, food or environmental sampling and facility inspections. Uncommon methods included industry consultation, tracer testing, anthropologic investigations and the use of food displays. Most outbreak investigations employed multiple methods to generate hypotheses and the context of the outbreak was an important determinant for some methods.
The multitude of hypothesis generation methods described and the use of multiple methods by most outbreak investigators point to the complexity of investigating enteric illness outbreaks. Many methods described are complementary with other methods or may be used in sequence as an investigation progress. For example, routine and enhanced surveillance questionnaires will often be collected before an outbreak is even identified, while hypothesis generating questionnaires are frequently used at the beginning of an outbreak when the focus of the investigation is quite broad. The use of descriptive epidemiology is generally based on questionnaire data and is often one of the first hypothesis generation methods employed in outbreak investigations. Other methods, such as food or environmental sampling, facility inspections and food handler testing may be used in conjunction with questionnaires, particularly if the outbreak occurred in one setting or at an event. Both open-ended and iterative interviewing frequently occur later in investigations when no obvious source has emerged or as new cases are identified.
Investigators consider many factors when choosing a hypothesis generation method. For example, the length of time that has elapsed between case exposure and the identification of outbreak impact investigation tools such as the collection of contaminated food and environmental samples or facility inspections and traceback investigations [ 43 – 45 ]. Cost and feasibility are also important considerations for many hypothesis generation methods. Analytic studies can be expensive and time consuming [ 46 ], while food and environmental sampling requires laboratory resources for testing [ 47 , 48 ]. Changes in method type used over time, for example increases in the use of anecdotal reports and purchase records, likely reflect the increase in available technology such as online reporting through social media, and availability of online records. The decline in the use of analytic methods may reflect the increased availability of other, less expensive, hypothesis generation methods such as population comparisons or purchase records.
Outbreak setting can impact the choice of hypothesis generation methods. Methods frequently used in single setting outbreaks include tailored menu-based interviewing, facility inspections and food handler testing. These methods are well-suited to these settings because the common connection across cases is obvious and the source is expected to be identified at a single location common to the cases, such as a restaurant or hospital. For outbreaks related to a single event such as weddings or conferences, analytic studies such as a retrospective cohort are well suited to investigating known exposed populations. In contrast, the use of purchase records, such as store loyalty cards or credit card statements, is utilised when the outbreak is among the general population and there appears to be no obvious connection between cases. Similarly, a review of existing information is a method used frequently in outbreaks among the general population when the range of plausible sources of illness is substantially larger than would be present in single event outbreaks. Outbreak setting thus has implications for the feasibility and usefulness of many hypothesis generation methods.
One finding of this scoping review is that hypothesis generation methods are not well reported within outbreak reports. Descriptions of hypothesis generation methods and sequence of events were often limited or entirely omitted from the publications. This incomplete reporting makes it difficult to interpret how frequently some methods are used by outbreak investigation teams compared to what outbreaks are written up and published in detail. Thus, it is likely that some common methods such as routine questionnaires were underreported and are thus underrepresented in this review. Methods that did not contribute to the identification of the source may also not be reported. Thorough reporting of all hypothesis generation methods used by outbreak investigators would allow for a more comprehensive understanding of the range and frequency of methods used to investigate outbreaks.
Most of the methods papers identified in this review focused on analytic studies, laboratory methods, traceback, interviews and questionnaires. No methods papers were identified related to several hypothesis generation methods reported in this review, including focus groups, iterative interviewing, open-ended interviewing, descriptive epidemiology, sub-cluster and outlier investigation, food or environmental sampling, facility inspections, food handler testing, review of existing information, menu or recipe analysis, anecdotal reports and social network analysis. The paucity of methods papers exploring hypothesis generation methods is an important literature gap. The relative merits of different hypothesis generation methods, their validity and reliability and comparable effectiveness across outbreak investigations, are needed to support outbreak investigator decision-making.
The frequencies of hypothesis generation methods reported in this scoping review may differ from their frequencies in practice as most outbreaks identified had successfully identified the source of the outbreak. Only 15% failed to identify the source of the outbreak, which is a much lower proportion than expected in practice [ 49 , 50 ]. This suggests that investigations where the source is not identified are less likely to be published and/or are published with few details, so they did not fulfil the inclusion criteria. This underreporting makes it impossible to accurately assess individual hypothesis generation methods' relative impact on investigation success based solely on published literature. Increased reporting of outbreak investigations where the source is not identified would improve our understanding of effective vs. ineffective hypothesis generation method use. Alternatively, organisations with access to administrative data on a full complement of outbreaks could analyse the relationship between the hypothesis generation methods used and associated outcomes of all outbreak investigations. For instance, Murphree et al . [ 49 ] compared the success of analytic studies to other methods in identifying a food vehicle across all outbreaks in the United States Foodborne Diseases Active Surveillance Network (FoodNet) catchment area. Analytic studies had a 47% success rate compared to all other methods with a 14% success rate [ 49 ], suggesting that analytic studies, where feasible, are more likely to lead to the identification of the source. However, given that analytic studies are not always feasible or appropriate, additional information on the relative success of other methods would help outbreak investigators choose appropriate methods to optimise the likelihood of successfully identifying the source. It would be valuable if outbreak investigators reported brief evaluations of their hypothesis generation methods to improve our understanding of the strengths and limitations of each method.
This review employed a comprehensive search strategy to identify enteric outbreak investigations and articles on hypothesis generation methods for outbreaks or other foodborne illness investigations. It is possible that despite our efforts some outbreak reports with hypothesis generation information were missed, as outbreaks are often not reported in the peer-reviewed literature and thus are not indexed in searchable bibliographic databases. To circumvent this shortfall, we performed a comprehensive grey literature search, however, it is possible some relevant reports were missed. It is also possible that there is some language bias, as the search was conducted in English and only papers reported in English or French were included in the review. This may have resulted in a failure of the search to identify relevant non-English papers. The effect of this on our results and conclusions is unknown. Lastly, because some methods identified in this review could be used for either hypothesis generation or hypothesis testing, we may have misclassified some uses of those methods as hypothesis generation when the investigators actually used the method for hypothesis testing. We relied on author reporting to understand when hypothesis generation was taking place, but incomplete or inadequate reporting may have resulted in misclassification that overestimated the extent to which some methods, such as analytic studies, are used to generate hypotheses.
This review demonstrated the range of hypothesis generation methods used in enteric illness outbreak investigations in humans. Most outbreaks were investigated using a combination of methods, highlighting the complexity of outbreak investigations and the requirement to have a suite of hypothesis generation approaches to choose from, as a single approach may not be appropriate in all situations. Research is needed to comprehensively understand the effectiveness of each hypothesis generation method in identifying the source of the outbreak, improving investigators' ability to choose the most suitable hypothesis generation methods to enable successful source identification.
Acknowledgements
The Public Health Agency of Canada library for their help in the procurement of publications. The Public Health Agency of Canada Centre for Food-borne, Environmental and Zoonotic Infectious Diseases, Outbreak Management Division contributors: Jennifer Cutler, Kristyn Franklin, Ashley Kerr, Vanessa Morton, Florence Tanguay, Joanne Tataryn, Kashmeera Meghnath, Mihaela Gheorghe, Shiona Glass-Kaastra.
Conflict of interest
Financial support.
This research received no specific grant from any funding agency, commercial or not-for-profit sectors.
Supplementary material
Information
- Author Services
Initiatives
You are accessing a machine-readable page. In order to be human-readable, please install an RSS reader.
All articles published by MDPI are made immediately available worldwide under an open access license. No special permission is required to reuse all or part of the article published by MDPI, including figures and tables. For articles published under an open access Creative Common CC BY license, any part of the article may be reused without permission provided that the original article is clearly cited. For more information, please refer to https://www.mdpi.com/openaccess .
Feature papers represent the most advanced research with significant potential for high impact in the field. A Feature Paper should be a substantial original Article that involves several techniques or approaches, provides an outlook for future research directions and describes possible research applications.
Feature papers are submitted upon individual invitation or recommendation by the scientific editors and must receive positive feedback from the reviewers.
Editor’s Choice articles are based on recommendations by the scientific editors of MDPI journals from around the world. Editors select a small number of articles recently published in the journal that they believe will be particularly interesting to readers, or important in the respective research area. The aim is to provide a snapshot of some of the most exciting work published in the various research areas of the journal.
Original Submission Date Received: .
- Active Journals
- Find a Journal
- Proceedings Series
- For Authors
- For Reviewers
- For Editors
- For Librarians
- For Publishers
- For Societies
- For Conference Organizers
- Open Access Policy
- Institutional Open Access Program
- Special Issues Guidelines
- Editorial Process
- Research and Publication Ethics
- Article Processing Charges
- Testimonials
- Preprints.org
- SciProfiles
- Encyclopedia

Article Menu

- Subscribe SciFeed
- Recommended Articles
- Author Biographies
- Google Scholar
- on Google Scholar
- Table of Contents
Find support for a specific problem in the support section of our website.
Please let us know what you think of our products and services.
Visit our dedicated information section to learn more about MDPI.
JSmol Viewer
Impacts of generative artificial intelligence in higher education: research trends and students’ perceptions.

1. Introduction
2. materials and methods.
- “Generative Artificial Intelligence” or “Generative AI” or “Gen AI”, AND;
- “Higher Education” or “University” or “College” or “Post-secondary”, AND;
- “Impact” or “Effect” or “Influence”.
- Q1— Does GenAI have more positive or negative effects on higher education? Options (to choose one): 1. It has more negative effects than positives; 2. It has more positive effects than negative; 3. There is a balance between positive and negative effects; 4. Don’t know.
- Q2— Identify the main positive effect of Gen AI in an academic context . Open-ended question.
- Q3— Identify the main negative effect of Gen AI in an academic context . Open-ended question.
3.1. Impacts of Gen AI in HE: Research Trends
3.1.1. he with gen ai, the key role that pedagogy must play, new ways to enhance the design and implementation of teaching and learning activities.
- Firstly, prompting in teaching should be prioritized as it plays a crucial role in developing students’ abilities. By providing appropriate prompts, educators can effectively guide students toward achieving their learning objectives.
- Secondly, configuring reverse prompting within the capabilities of Gen AI chatbots can greatly assist students in monitoring their learning progress. This feature empowers students to take ownership of their education and fosters a sense of responsibility.
- Furthermore, it is essential to embed digital literacy in all teaching and learning activities that aim to leverage the potential of the new Gen AI assistants. By equipping students with the necessary skills to navigate and critically evaluate digital resources, educators can ensure that they are prepared for the digital age.
The Student’s Role in the Learning Experience
The key teacher’s role in the teaching and learning experience, 3.1.2. assessment in gen ai/chatgpt times, the need for new assessment procedures, 3.1.3. new challenges to academic integrity policies, new meanings and frontiers of misconduct, personal data usurpation and cheating, 3.2. students’ perceptions about the impacts of gen ai in he.
- “It harms the learning process”: ▪ “What is generated by Gen AI has errors”; ▪ “Generates dependence and encourages laziness”; ▪ “Decreases active effort and involvement in the learning/critical thinking process”.
4. Discussion
- Training: providing training for both students and teachers on effectively using and integrating Gen AI technologies into teaching and learning practices.
- Ethical use and risk management: developing policies and guidelines for ethical use and risk management associated with Gen AI technologies.
- Incorporating AI without replacing humans: incorporating AI technologies as supplementary tools to assist teachers and students rather than replacements for human interaction.
- Continuously enhancing holistic competencies: encouraging the use of AI technologies to enhance specific skills, such as digital competence and time management, while ensuring that students continue to develop vital transferable skills.
- Fostering a transparent AI environment: promoting an environment in which students and teachers can openly discuss the benefits and concerns associated with using AI technologies.
- Data privacy and security: ensuring data privacy and security using AI technologies.
- The dynamics of technological support to align with the most suitable Gen AI resources;
- The training policy to ensure that teachers, students, and academic staff are properly trained to utilize the potential of Gen AI and its tools;
- Security and data protection policies;
- Quality and ethical action policies.
5. Conclusions
- Database constraints: the analysis is based on existing publications in SCOPUS and the Web of Science, potentially omitting relevant research from other sources.
- Inclusion criteria: due to the early stage of scientific production on this topic, all publications were included in the analysis, rather than focusing solely on articles from highly indexed journals and/or with a high number of citations as recommended by bibliometric and systematic review best practices.
- Dynamic landscape: the rate of publications on Gen AI has been rapidly increasing and diversifying in 2024, highlighting the need for ongoing analysis to track trends and changes in scientific thinking.
Author Contributions
Institutional review board statement, informed consent statement, data availability statement, conflicts of interest.
- Akakpo, Martin Gameli. 2023. Skilled for the Future: Information Literacy for AI Use by University Students in Africa and the Role of Librarians. Internet Reference Services Quarterly 28: 19–26. [ Google Scholar ] [ CrossRef ]
- AlAfnan, Mohammad Awad, Samira Dishari, Marina Jovic, and Koba Lomidze. 2023. ChatGPT as an Educational Tool: Opportunities, Challenges, and Recommendations for Communication, Business Writing, and Composition Courses. Journal of Artificial Intelligence and Technology 3: 60–68. [ Google Scholar ] [ CrossRef ]
- Almaraz-López, Cristina, Fernando Almaraz-Menéndez, and Carmen López-Esteban. 2023. Comparative Study of the Attitudes and Perceptions of University Students in Business Administration and Management and in Education toward Artificial Intelligence. Education Sciences 13: 609. [ Google Scholar ] [ CrossRef ]
- Al-Zahrani, Abdulrahman. 2023. The impact of generative AI tools on researchers and research: Implications for academia in higher education. Innovations in Education and Teaching International , 1–15. [ Google Scholar ] [ CrossRef ]
- Athilingam, Ponrathi, and Hong-Gu He. 2023. ChatGPT in nursing education: Opportunities and challenges. Teaching and Learning in Nursing 19: 97–101. [ Google Scholar ] [ CrossRef ]
- Álvarez-Álvarez, Carmen, and Samuel Falcon. 2023. Students’ preferences with university teaching practices: Analysis of testimonials with artificial intelligence. Educational Technology Research and Development 71: 1709–24. [ Google Scholar ] [ CrossRef ]
- Bannister, Peter, Elena Alcalde Peñalver, and Alexandra Santamaría Urbieta. 2023. Transnational higher education cultures and generative AI: A nominal group study for policy development in English medium instruction. Journal for Multicultural Education . ahead-of-print . [ Google Scholar ] [ CrossRef ]
- Bearman, Margaret, and Rola Ajjawi. 2023. Learning to work with the black box: Pedagogy for a world with artificial intelligence. British Journal of Educational Technology 54: 1160–73. [ Google Scholar ] [ CrossRef ]
- Boháček, Matyas. 2023. The Unseen A+ Student: Evaluating the Performance and Detectability of Large Language Models in the Classroom. CEUR Workshop Proceedings 3487: 89–100. Available online: https://openreview.net/pdf?id=9ZKJLYg5EQ (accessed on 7 January 2024).
- Chan, Cecilia Ka Yuk. 2023. A comprehensive AI policy education framework for university teaching and learning. International Journal of Educational Technology in Higher Education 20: 38. [ Google Scholar ] [ CrossRef ]
- Chan, Cecilia Ka Yuk, and Wenjie Hu. 2023. Students’ voices on generative AI: Perceptions, benefits, and challenges in higher education. International Journal of Educational Technology in Higher Education 20: 43. [ Google Scholar ] [ CrossRef ]
- Chan, Cecilia Ka Yuk, and Wenxin Zhou. 2023. An expectancy value theory (EVT) based instrument for measuring student perceptions of generative AI. Smart Learning Environments 10: 64. [ Google Scholar ] [ CrossRef ]
- Chang, Daniel H., Michael Pin-Chuan Lin, Shiva Hajian, and Quincy Q. Wang. 2023. Educational Design Principles of Using AI Chatbot That Supports Self-Regulated Learning in Education: Goal Setting, Feedback, and Personalization. Sustainability 15: 12921. [ Google Scholar ] [ CrossRef ]
- Chiu, Thomas. 2023. The impact of Generative AI (GenAI) on practices, policies and research direction in education: A case of ChatGPT and Midjourney. Interactive Learning Environments , 1–17. [ Google Scholar ] [ CrossRef ]
- Chun, John, and Katherine Elkins. 2023. The Crisis of Artificial Intelligence: A New Digital Humanities Curriculum for Human-Centred AI. International Journal of Humanities and Arts Computing 17: 147–67. [ Google Scholar ] [ CrossRef ]
- Cowling, Michael, Joseph Crawford, Kelly-Ann Allen, and Michael Wehmeyer. 2023. Using leadership to leverage ChatGPT and artificial intelligence for undergraduate and postgraduate research supervision. Australasian Journal of Educational Technology 39: 89–103. [ Google Scholar ] [ CrossRef ]
- Crawford, Joseph, Carmen Vallis, Jianhua Yang, Rachel Fitzgerald, Christine O’Dea, and Michael Cowling. 2023a. Editorial: Artificial Intelligence is Awesome, but Good Teaching Should Always Come First. Journal of University Teaching & Learning Practice 20: 01. [ Google Scholar ] [ CrossRef ]
- Crawford, Joseph, Michael Cowling, and Kelly-Ann Allen. 2023b. Leadership is needed for ethical ChatGPT: Character, assessment, and learning using artificial intelligence (AI). Journal of University Teaching & Learning Practice 20: 02. [ Google Scholar ] [ CrossRef ]
- Currie, Geoffrey. 2023a. A Conversation with ChatGPT. Journal of Nuclear Medicine Technology 51: 255–60. [ Google Scholar ] [ CrossRef ] [ PubMed ]
- Currie, Geoffrey. 2023b. GPT-4 in Nuclear Medicine Education: Does It Outperform GPT-3.5? Journal of Nuclear Medicine Technology 51: 314–17. [ Google Scholar ] [ CrossRef ] [ PubMed ]
- Currie, Geoffrey, and Kym Barry. 2023. ChatGPT in Nuclear Medicine Education. Journal of Nuclear Medicine Technology 51: 247–54. [ Google Scholar ] [ CrossRef ]
- Currie, Geoffrey, Clare Singh, Tarni Nelson, Caroline Nabasenja, Yazan Al-Hayek, and Kelly Spuur. 2023. ChatGPT in medical imaging higher education. Radiography 29: 792–99. [ Google Scholar ] [ CrossRef ]
- Dai, Yun, Ang Liu, and Cher P. Lim. 2023. Reconceptualizing Chatgpt and Generative AI as a Student-driven Innovation in Higher Education. Procedia CIRP Volume 119: 84–90. [ Google Scholar ] [ CrossRef ]
- Dogru, Tarik, Nathana Line, Lydia Hanks, Fulya Acikgoz, Je’Anna Abbott, Selim Bakir, Adiyukh Berbekova, Anil Bilgihan, Ali Iskender, Murat Kizildag, and et al. 2023. The implications of generative artificial intelligence in academic research and higher education in tourism and hospitality. Tourism Economics 30: 1083–94. [ Google Scholar ] [ CrossRef ]
- Duong, Cong Doanh, Trong Nghia Vu, and Thi Viet Nga Ngo. 2023. Applying a modified technology acceptance model to explain higher education students’ usage of ChatGPT: A serial multiple mediation model with knowledge sharing as a moderator. The International Journal of Management Education 21: 100883. [ Google Scholar ] [ CrossRef ]
- Eager, Bronwyn, and Ryan Brunton. 2023. Prompting Higher Education Towards AI-Augmented Teaching and Learning Practice. Journal of University Teaching & Learning Practice 20: 5. [ Google Scholar ] [ CrossRef ]
- Elkhodr, Mahmoud, Ergun Gide, Robert Wu, and Omar Darwish. 2023. ICT students’ perceptions towards ChatGPT: An experimental reflective lab analysis. STEM Education 3: 70–88. [ Google Scholar ] [ CrossRef ]
- Farrelly, Tom, and Nick Baker. 2023. Generative Artificial Intelligence: Implications and Considerations for Higher Education Practice. Education Sciences 13: 1109. [ Google Scholar ] [ CrossRef ]
- Farrokhnia, Mohammadreza, Seyyed Banihashem, Seyyed Kazem Banihashem, Omid Noroozi, and Arjen Wals. 2023. A SWOT analysis of ChatGPT: Implications for educational practice and research. Innovations in Education and Teaching International 61: 460–74. [ Google Scholar ] [ CrossRef ]
- Gong, Furong. 2023. The Impact of Generative AI like ChatGPT on Digital Literacy Education in University Libraries. Documentation, Information & Knowledge 40: 97–106, 156. [ Google Scholar ] [ CrossRef ]
- Han, Bingyi, Sadia Nawaz, George Buchanan, and Dana McKay. 2023. Ethical and Pedagogical Impacts of AI in Education. In Artificial Intelligence in Education . Edited by Ning Wang, Genaro Rebolledo-Mendez, Noboru Matsuda, Olga Santos and Vania Dimitrova. Lecture Notes in Computer Science. Cham: Springer, pp. 667–73. [ Google Scholar ] [ CrossRef ]
- Hassoulas, Athanasios, Ned Powell, Lindsay Roberts, Katja Umla-Runge, Laurence Gray, and Marcus J. Coffey. 2023. Investigating marker accuracy in differentiating between university scripts written by students and those produced using ChatGPT. Journal of Applied Learning and Teaching 6: 71–77. [ Google Scholar ] [ CrossRef ]
- Hernández-Leo, Davinia. 2023. ChatGPT and Generative AI in Higher Education: User-Centered Perspectives and Implications for Learning Analytics. CEUR Workshop Proceedings , 1–6. Available online: https://ceur-ws.org/Vol-3542/paper2.pdf (accessed on 7 January 2024).
- Hidayat-ur-Rehman, Imdadullah, and Yasser Ibrahim. 2023. Exploring factors influencing educators’ adoption of ChatGPT: A mixed method approach. Interactive Technology and Smart Education . ahead-of-print . [ Google Scholar ] [ CrossRef ]
- Ilieva, Galina, Tania Yankova, Stanislava Klisarova-Belcheva, Angel Dimitrov, Marin Bratkov, and Delian Angelov. 2023. Effects of Generative Chatbots in Higher Education. Information 14: 492. [ Google Scholar ] [ CrossRef ]
- Javaid, Mohd, Abid Haleem, Ravi Pratap Singh, Shahbaz Khan, and Haleem Ibrahim. 2023. Unlocking the opportunities through ChatGPT Tool towards ameliorating the education system. Bench Council Transactions on Benchmarks, Standards and Evaluations 3: 100115. [ Google Scholar ] [ CrossRef ]
- Kaplan-Rakowski, Regina, Kimberly Grotewold, Peggy Hartwick, and Kevin Papin. 2023. Generative AI and Teachers’ Perspectives on Its Implementation in Education. Journal of Interactive Learning Research 34: 313–38. Available online: https://www.learntechlib.org/primary/p/222363/ (accessed on 7 January 2024).
- Karunaratne, Thashmee, and Adenike Adesina. 2023. Is it the new Google: Impact of ChatGPT on Students’ Information Search Habits. Paper presented at the European Conference on e-Learning (ECEL 2023), Pretoria, South Africa, October 26–27; pp. 147–55. [ Google Scholar ] [ CrossRef ]
- Kelly, Andrew, Miriam Sullivan, and Katrina Strampel. 2023. Generative artificial intelligence: University student awareness, experience, and confidence in use across disciplines. Journal of University Teaching & Learning Practice 20: 12. [ Google Scholar ] [ CrossRef ]
- Kohnke, Lucas, Benjamin Luke Moorhouse, and Di Zou. 2023. Exploring generative artificial intelligence preparedness among university language instructors: A case study. Computers and Education: Artificial Intelligence 5: 100156. [ Google Scholar ] [ CrossRef ]
- Laker, Lauren, and Mark Sena. 2023. Accuracy and detection of student use of ChatGPT in business analytics courses. Issues in Information Systems 24: 153–63. [ Google Scholar ] [ CrossRef ]
- Lemke, Claudia, Kathrin Kirchner, Liadan Anandarajah, and Florian Herfurth. 2023. Exploring the Student Perspective: Assessing Technology Readiness and Acceptance for Adopting Large Language Models in Higher Education. Paper presented at the European Conference on e-Learning, (ECEL 2023), Pretoria, South Africa, October 26–27; pp. 156–64. [ Google Scholar ] [ CrossRef ]
- Limna, Pongsakorn, Tanpat Kraiwanit, Kris Jangjarat, and Prapasiri Klayklung. 2023a. The use of ChatGPT in the digital era: Perspectives on chatbot implementation. Journal of Applied Learning and Teaching 6: 64–74. [ Google Scholar ] [ CrossRef ]
- Limna, Pongsakorn, Tanpat Kraiwanit, Kris Jangjarat, and Yarnaphat Shaengchart. 2023b. Applying ChatGPT as a new business strategy: A great power comes with great responsibility [Special issue]. Corporate & Business Strategy Review 4: 218–26. [ Google Scholar ] [ CrossRef ]
- Lopezosa, Carlos, Carles Lluís Codina, Carles Pont-Sorribes, and Mari Vállez. 2023. Use of Generative Artificial Intelligence in the Training of Journalists: Challenges, Uses and Training Proposal. Profesional De La información Information Professional 32: 1–12. [ Google Scholar ] [ CrossRef ]
- Martineau, Kim. 2023. What Is Generative AI? IBM Research Blog . April 20. Available online: https://research.ibm.com/blog/what-is-generative-AI (accessed on 7 January 2024).
- Mondal, Himel, Shaikat Mondal, and Indrashis Podder. 2023. Using ChatGPT for Writing Articles for Patients’ Education for Dermatological Diseases: A Pilot Study. Indian Dermatology Online Journal 14: 482–86. [ Google Scholar ] [ CrossRef ]
- Moorhouse, Benjamin, Marie Alina Wan, and Yuwei Wan. 2023. Generative AI tools and assessment: Guidelines of the world’s top-ranking universities. Computers and Education Open 5: 100151. [ Google Scholar ] [ CrossRef ]
- Overono, Acacia L., and Annie Ditta. 2023. The Rise of Artificial Intelligence: A Clarion Call for Higher Education to Redefine Learning and Reimagine Assessment. College Teaching , 1–4. [ Google Scholar ] [ CrossRef ]
- Page, Matthew J., Joanne E. McKenzie, Patrick M. Bossuyt, Isabelle Boutron, Tammy C. Hoffmann, Cynthia D. Mulrow, Larissa Shamseer, Jennifer M. Tetzlaff, Elie A. Akl, Sue E. Brennan, and et al. 2021. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. BMJ 372: n71. [ Google Scholar ] [ CrossRef ]
- Pechenkina, Ekaterina. 2023. Artificial intelligence for good? Challenges and possibilities of AI in higher education from a data justice perspective. In Higher Education for Good: Teaching and Learning Futures . Edited by Laura Czerniewicz and Catherine Cronin. Cambridge, UK: Open Book Publishers, pp. 239–66. [ Google Scholar ] [ CrossRef ]
- Perkins, Mike, Jasper Roe, Darius Postma, James McGaughran, and Don Hickerson. 2023. Detection of GPT-4 Generated Text in Higher Education: Combining Academic Judgement and Software to Identify Generative AI Tool Misuse. Journal of Academic Ethics 22: 89–113. [ Google Scholar ] [ CrossRef ]
- Pitso, Teboho. 2023. Post-COVID-19 Higher Learning: Towards Telagogy, A Web-Based Learning Experience. IAFOR Journal of Education 11: 39–59. [ Google Scholar ] [ CrossRef ]
- Plata, Sterling, Maria Ana De Guzman, and Arthea Quesada. 2023. Emerging Research and Policy Themes on Academic Integrity in the Age of Chat GPT and Generative AI. Asian Journal of University Education 19: 743–58. [ Google Scholar ] [ CrossRef ]
- Rudolph, Jürgen, Samson Tan, and Shannon Tan. 2023a. War of the chatbots: Bard, Bing Chat, ChatGPT, Ernie and beyond. The new AI gold rush and its impact on higher education. Journal of Applied Learning and Teaching 6: 364–89. [ Google Scholar ] [ CrossRef ]
- Rudolph, Jürgen, Samson Tan, and Shannon Tan. 2023b. ChatGPT: Bullshit spewer or the end of traditional assessments in higher education? Journal of Applied Learning and Teaching 6: 342–63. [ Google Scholar ] [ CrossRef ]
- Ryall, Adelle, and Stephen Abblitt. 2023. “A Co-Pilot for Learning Design?”: Perspectives from Learning Designers on the Uses, Challenges, and Risks of Generative Artificial Intelligence in Higher Education. In People, Partnerships and Pedagogies. Proceedings ASCILITE 2023 . Edited by Thomas Cochrane, Vickel Narayan, Cheryl Brown, MacCallum Kathryn, Elisa Bone, Christopher Deneen, Robert Vanderburg and Brad Hurren. Christchurch: Te Pae Conference Center, pp. 525–30. [ Google Scholar ] [ CrossRef ]
- Santiago, Cereneo S., Steve I. Embang, Ricky B. Acanto, Kem Warren P. Ambojia, Maico Demi B. Aperocho, Benedicto B. Balilo, Erwin L. Cahapin, Marjohn Thomas N. Conlu, Samson M. Lausa, Ester Y. Laput, and et al. 2023. Utilization of Writing Assistance Tools in Research in Selected Higher Learning Institutions in the Philippines: A Text Mining Analysis. International Journal of Learning, Teaching and Educational Research 22: 259–84. [ Google Scholar ] [ CrossRef ]
- Solopova, Veronika, Eiad Rostom, Fritz Cremer, Adrian Gruszczynski, Sascha Witte, Chengming Zhang, Fernando Ramos López, Lea Plößl, Florian Hofmann, Ralf Romeike, and et al. 2023. PapagAI: Automated Feedback for Reflective Essays. In KI 2023: Advances in Artificial Intelligence. KI 2023 . Lecture Notes in Computer Science (Including Subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics). Cham: Springer, vol. 14236, pp. 198–206. [ Google Scholar ] [ CrossRef ]
- Sridhar, Pragnya, Aidan Doyle, Arav Agarwal, Christopher Bogart, Jaromir Savelka, and Majd Sakr. 2023. Harnessing LLMs in Curricular Design: Using GPT-4 to Support Authoring of Learning Objectives. CEUR Workshop Proceedings 3487: 139–50. [ Google Scholar ]
- Sullivan, Miriam, Andrew Kelly, and Paul McLaughlan. 2023. ChatGPT in higher education: Considerations for academic integrity and student learning. Journal of Applied Learning and Teaching 6: 31–40. [ Google Scholar ] [ CrossRef ]
- Tominc, Polona, and Maja Rožman. 2023. Artificial Intelligence and Business Studies: Study Cycle Differences Regarding the Perceptions of the Key Future Competences. Education Sciences 13: 580. [ Google Scholar ] [ CrossRef ]
- van den Berg, Geesje, and Elize du Plessis. 2023. ChatGPT and Generative AI: Possibilities for Its Contribution to Lesson Planning, Critical Thinking and Openness in Teacher Education. Education Sciences 13: 998. [ Google Scholar ] [ CrossRef ]
- Walczak, Krzysztof, and Wojciech Cellary. 2023. Challenges for higher education in the era of widespread access to Generative AI. Economics and Business Review 9: 71–100. [ Google Scholar ] [ CrossRef ]
- Wang, Ting, Brady D. Lund, Agostino Marengo, Alessandro Pagano, Nishith Reddy Mannuru, Zoë A. Teel, and Jenny Pange. 2023. Exploring the Potential Impact of Artificial Intelligence (AI) on International Students in Higher Education: Generative AI, Chatbots, Analytics, and International Student Success. Applied Sciences 13: 6716. [ Google Scholar ] [ CrossRef ]
- Watermeyer, Richard, Lawrie Phipps, Donna Lanclos, and Cathryn Knight. 2023. Generative AI and the Automating of Academia. Postdigital Science and Education 6: 446–66. [ Google Scholar ] [ CrossRef ]
- Wolf, Leigh, Tom Farrelly, Orna Farrell, and Fiona Concannon. 2023. Reflections on a Collective Creative Experiment with GenAI: Exploring the Boundaries of What is Possible. Irish Journal of Technology Enhanced Learning 7: 1–7. [ Google Scholar ] [ CrossRef ]
- Yilmaz, Ramazan, and Fatma Gizem Karaoglan Yilmaz. 2023. The effect of generative artificial intelligence (AI)-based tool use on students’ computational thinking skills, programming self-efficacy and motivation. Computers and Education: Artificial Intelligence 4: 100147. [ Google Scholar ] [ CrossRef ]
- Zawiah, Mohammed, Fahmi Y. Al-Ashwal, Lobna Gharaibeh, Rana Abu Farha, Karem H. Alzoubi, Khawla Abu Hammour, Qutaiba A. Qasim, and Fahd Abrah. 2023. ChatGPT and Clinical Training: Perception, Concerns, and Practice of Pharm-D Students. Journal of Multidisciplinary Healthcare 16: 4099–110. [ Google Scholar ] [ CrossRef ]
Click here to enlarge figure
| Selected Group of Students | Students Who Answered the Questionnaire | |||
|---|---|---|---|---|
| M | F | M | F | |
| 1st year | 59 | 5 | 34 | 2 |
| 2nd year | 36 | 5 | 29 | 4 |
| 1st year | 39 | 3 | 24 | 2 |
| 2nd year | 21 | 2 | 15 | 2 |
| Country | N. | Country | N. | Country | N. | Country | N. |
|---|---|---|---|---|---|---|---|
| Australia | 16 | Italy | 2 | Egypt | 1 | South Korea | 1 |
| United States | 7 | Saudi Arabia | 2 | Ghana | 1 | Sweden | 1 |
| Singapore | 5 | South Africa | 2 | Greece | 1 | Turkey | 1 |
| Hong Kong | 4 | Thailand | 2 | India | 1 | United Arab Emirates | 1 |
| Spain | 4 | Viet Nam | 2 | Iraq | 1 | Yemen | 1 |
| United Kingdom | 4 | Bulgaria | 1 | Jordan | 1 | ||
| Canada | 3 | Chile | 1 | Malaysia | 1 | ||
| Philippines | 3 | China | 1 | Mexico | 1 | ||
| Germany | 2 | Czech Republic | 1 | New Zealand | 1 | ||
| Ireland | 2 | Denmark | 1 | Poland | 1 |
| Country | N. | Country | N. | Country | N. | Country | N. |
|---|---|---|---|---|---|---|---|
| Singapore | 271 | United States | 15 | India | 2 | Iraq | 0 |
| Australia | 187 | Italy | 11 | Turkey | 2 | Jordan | 0 |
| Hong Kong | 37 | United Kingdom | 6 | Denmark | 1 | Poland | 0 |
| Thailand | 33 | Canada | 6 | Greece | 1 | United Arab Emirates | 0 |
| Philippines | 31 | Ireland | 6 | Sweden | 1 | Yemen | 0 |
| Viet Nam | 29 | Spain | 6 | Saudi Arabia | 1 | ||
| Malaysia | 29 | South Africa | 6 | Bulgaria | 1 | ||
| South Korea | 29 | Mexico | 3 | Czech Republic | 0 | ||
| China | 17 | Chile | 3 | Egypt | 0 | ||
| New Zealand | 17 | Germany | 2 | Ghana | 0 |
| Categories | Subcategories | Nr. of Documents | References |
|---|---|---|---|
| HE with Gen AI | 15 | ( ); ( ); ( ); ( ); ( ); ( ); ( ); ( ); ( ); ( ); ( ); ( ); ( ); ( ); ( ); ( ); ( ); ( ). | |
| 15 | ( ); ( ); ( ); ( ); ( ); ( ); ( ); ( ); ( ); ( ); ( ); ( ); ( ); ( ); ( ); ( ). | ||
| 14 | ( ); ( ); ( ); ( ); ( ); ( ); ( ); ( ); ( ); ( ); ( ); ( ); ( ); ( ). | ||
| 8 | ( ); ( ); ( ); ( ); ( ); ( ); ( ); ( ). | ||
| Assessment in Gen AI/ChatGPT times | 8 | ( ); ( ); ( ); ( ); ( ); ( ); ( ); ( ). | |
| New challenges to academic integrity policies | 4 | ( ); ( ); ( ); ( ). |
| Have You Tried Using a Gen AI Tool? | Nr. | % |
|---|---|---|
| Yes | 52 | 46.4% |
| No | 60 | 53.6% |
| Categories and Subcategories | % | Unit of Analysis (Some Examples) |
|---|---|---|
| 1. Learning support: | ||
| 1.1. Helpful to solve doubts, to correct errors | 34.6% | |
| 1.2. Helpful for more autonomous and self-regulated learning | 19.2% | |
| 2. Helpful to carry out the academic assignments/individual or group activities | 17.3% | |
| 3. Facilitates research/search processes | ||
| 3.1. Reduces the time spent with research | 13.5% | |
| 3.2. Makes access to information easier | 9.6% | |
| 4. Reduction in teachers’ workload | 3.9% | |
| 5. Enables new teaching methods | 1.9% | |
| Categories and Subcategories | % | Unit of Analysis (Some Examples) |
|---|---|---|
| 1. Harms the learning process: | ||
| 1.1. What is generated by Gen AI has errors | 13.5% | |
| 1.2. Generates dependence and encourages laziness | 15.4% | |
| 1.3. Decreases active effort and involvement in the learning/critical thinking process | 28.8% | |
| 2. Encourages plagiarism and incorrect assessment procedures | 17.3% | |
| 3. Reduces relationships with teachers and interpersonal relationships | 9.6% | |
| 4. No negative effect—as it will be necessary to have knowledge for its correct use | 7.7% | |
| 5. Don’t know | 7.7% | |
| The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
Share and Cite
Saúde, S.; Barros, J.P.; Almeida, I. Impacts of Generative Artificial Intelligence in Higher Education: Research Trends and Students’ Perceptions. Soc. Sci. 2024 , 13 , 410. https://doi.org/10.3390/socsci13080410
Saúde S, Barros JP, Almeida I. Impacts of Generative Artificial Intelligence in Higher Education: Research Trends and Students’ Perceptions. Social Sciences . 2024; 13(8):410. https://doi.org/10.3390/socsci13080410
Saúde, Sandra, João Paulo Barros, and Inês Almeida. 2024. "Impacts of Generative Artificial Intelligence in Higher Education: Research Trends and Students’ Perceptions" Social Sciences 13, no. 8: 410. https://doi.org/10.3390/socsci13080410
Article Metrics
Article access statistics, further information, mdpi initiatives, follow mdpi.

Subscribe to receive issue release notifications and newsletters from MDPI journals

IMAGES
VIDEO
COMMENTS
Hypothesis generation is an early and critical step in any hypothesis-driven clinical research project. Because it is not yet a well-understood cognitive process, the need to improve the process goes unrecognized. Without an impactful hypothesis, the significance of any research project can be questionable, regardless of the rigor or diligence applied in other steps of the study, e.g., study ...
Thus, hypothesis generation is an important initial step in the research workflow, reflecting accumulating evidence and experts' stance. In this article, we overview the genesis and importance of scientific hypotheses and their relevance in the era of the coronavirus disease 2019 (COVID-19) pandemic. ... Review articles are generated by ...
However, scientific hypothesis generation with human participants is rare in the literature. Although hypothesis generation is an early step in scientific studies and research projects 1 and its critical role has been broadly recognized 27-29, few studies have focused on understanding the principles or exploring the mechanisms of the process.
A review for clinical outcomes research: hypothesis generation, data strategy, and hypothesis-driven statistical analysis. ... The days of relying upon the "chart review" for definitive answers has passed us by. How, then, can we answer important clinical questions using current tools from the rapidly developing world of outcomes research ...
Hypothesis Generation is a literature-based discovery approach that utilizes existing literature to automatically generate implicit biomedical associations and provide reasonable predictions for future research. ... Ulrich Dirnagl, 2021. Improving target assessment in biomedical research: the GOT-IT recommendations. Nature reviews Drug ...
The hypothesis-generating mode of research has been primarily practiced in basic science but has recently been extended to clinical-translational work as well. Just as in basic science, this approach to research can facilitate insights into human health and disease mechanisms and provide the crucially needed data set of the full spectrum of ...
Automated hypothesis generation (HG) focuses on uncovering hidden connections within the extensive information that is publicly available. This domain has become increasingly popular, thanks to modern machine learning algorithms. However, the automated evaluation of HG systems is still an open problem, especially on a larger scale. This paper presents a novel benchmarking framework Dyport for ...
the scientific hypothesis to determine the answer to research questions 2,4. Scientific hypothesis generation and scientific hypothesis testing are distinct processes 2,5. In clinical research, research questions are often delineated without the support of systematic data analysis (i.e., not data-driven) 2,6,7. Using and analyzing existing data ...
A review for clinical outcomes research: hypothesis generation, data strategy, and hypothesis-driven statistical analysis Surg Endosc. 2011 Jul;25(7):2254-60. doi: 10.1007/s00464-010-1543-7. Epub 2011 Feb 27. Authors David C Chang 1 , Mark A Talamini. Affiliation 1 Department of Surgery ...
Background Scientific hypothesis generation is a critical step in scientific research that determines the direction and impact of any investigation. Despite its vital role, we have limited knowledge of the process itself, hindering our ability to address some critical questions. Objective To what extent can secondary data analytic tools facilitate scientific hypothesis generation during ...
The hypothesis-generating mode of research has been primarily practiced in basic science but has recently been extended to clinical-translational work as well. Just as in basic science, this approach to research can facilitate insights into human health and disease mechanisms and provide the crucially needed data set of the full spectrum of ...
In science, experimentation and hypothesis generation often form an iterative cycle: a researcher asks a question, collects data and adjusts the question or asks a fresh one.
Principle 1 suggests that hypothesis-generation processes are a general case of cued recall in that the data or symptoms observed cue the retrieval of diagnostic hypotheses from either episodic long-term memory or knowledge. Note, however, that the retrieval goals in a hypothesis-generation task differ from the retrieval goals in the typical ...
The proposed hypothesis generation framework (HGF) finds "crisp semantic associations" among entities of interest - that is a step towards bridging such gaps. ... A manual review of the literature is performed to find evidences for some of the associations found only by the HGF; Table 2 summarizes these results.
The first part of the article demonstrates that as long as hypotheses are sparse (i.e., index less than half of the possible entities in the domain) then a positive test strategy is near optimal. The second part of this article then demonstrates that a preference for sparse hypotheses (a sparsity bias) emerges as a natural consequence of the ...
While hypothesis testing is a highly formalized activity, hypothesis generation remains largely informal. We propose a systematic procedure to generate novel hypotheses about human behavior, which uses the capacity of machine learning algorithms to notice patterns people might not. We illustrate the procedure with a concrete application: judge ...
Study flow and data sets used. The 2 × 2 study compared the hypothesis generation process of the clinical researchers with and without VIADS on the same datasets (), with the same study scripts (), and within the same timeframe (2 hours/study session), and they all followed the think-aloud method.The participants were separated into experienced and inexperienced clinical researchers based on ...
Jens Ludwig & Sendhil Mullainathan, 2024. "Machine Learning as a Tool for Hypothesis Generation," The Quarterly Journal of Economics, vol 139 (2), pages 751-827. Founded in 1920, the NBER is a private, non-profit, non-partisan organization dedicated to conducting economic research and to disseminating research findings among academics, public ...
The first part of the article demonstrates that as long as hypotheses are sparse (i.e., index less than half of the possible entities in the domain) then a positive test strategy is near optimal. The second part of this article then demonstrates that a preference for sparse hypotheses (a sparsity bias) emerges as a natural consequence of the ...
We champion the potential of AI for neuroscience exploration. We highlight both implicit, 'uninterpretable' models as aids in hypothesis formulation and symbolic regression for explicit hypothesis generation. For researchers from non-neuroscience backgrounds, we discuss domain-specific considerations in integrating AI into neuroscience research.
This is the essence of hypothesis generation. A hypothesis emerges from a set of underlying assumptions. It is an articulation of how those assumptions are expected to play out in a given context. In short, a hypothesis is an intelligent, articulated guess that is the basis for taking action and assessing outcomes.
Hypothesis Generation. Economics should be, as a science, concerned with formulating theories of ideas and reality that produce descriptions of how to understand phenomenon and create experiences, hypotheses generation, and data which need to be proven or disproven through testing and further analyses. ... In their review of agent-based ...
Foundational Models (FMs) are emerging as the cornerstone of the biomedical AI ecosystem due to their ability to represent and contextualize multimodal biomedical data. These capabilities allow FMs to be adapted for various tasks, including biomedical reasoning, hypothesis generation, and clinical decision-making. This review paper examines the foundational components of an ethical and ...
Literature research, vital for scientific advancement, is overwhelmed by the vast ocean of available information. Addressing this, we propose an automated review generation method based on Large Language Models (LLMs) to streamline literature processing and reduce cognitive load. In case study on propane dehydrogenation (PDH) catalysts, our method swiftly generated comprehensive reviews from ...
The eigenstate thermalization hypothesis describes how isolated many-body quantum systems reach thermal equilibrium. However, quantum many-body scars and Hilbert space fragmentation violate this hypothesis and cause nonthermal behavior. We demonstrate that Hilbert space fragmentation may arise from lattice geometry in a spin-$\\frac{1}{2}$ model that conserves the number of domain walls. We ...
This review demonstrated the range of hypothesis generation methods used in enteric illness outbreak investigations in humans. Most outbreaks were investigated using a combination of methods, highlighting the complexity of outbreak investigations and the requirement to have a suite of hypothesis generation approaches to choose from, as a single ...
In this paper, the effects of the rapid advancement of generative artificial intelligence (Gen AI) in higher education (HE) are discussed. A mixed exploratory research approach was employed to understand these impacts, combining analysis of current research trends and students' perceptions of the effects of Gen AI tools in academia. Through bibliometric analysis and systematic literature ...